Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
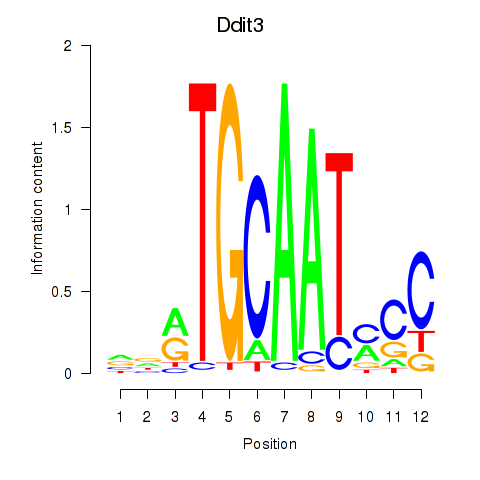
Results for Ddit3
Z-value: 0.69
Transcription factors associated with Ddit3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ddit3
|
ENSRNOG00000006789 | DNA-damage inducible transcript 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ddit3 | rn6_v1_chr7_+_70580198_70580252 | -0.49 | 4.0e-01 | Click! |
Activity profile of Ddit3 motif
Sorted Z-values of Ddit3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_6031655 | 0.32 |
ENSRNOT00000092016
|
AABR07042733.2
|
|
| chr15_+_64939527 | 0.29 |
ENSRNOT00000045865
|
AABR07018556.1
|
|
| chr16_-_49820235 | 0.28 |
ENSRNOT00000029628
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr1_+_168668707 | 0.26 |
ENSRNOT00000015108
|
AC107531.3
|
|
| chrX_+_137934484 | 0.25 |
ENSRNOT00000049046
|
LOC317588
|
hypothetical protein LOC317588 |
| chr2_-_112368557 | 0.25 |
ENSRNOT00000075017
|
AABR07009787.1
|
|
| chr17_+_48436720 | 0.25 |
ENSRNOT00000074309
|
AABR07027902.1
|
|
| chr1_-_4560789 | 0.25 |
ENSRNOT00000082415
|
Adgb
|
androglobin |
| chr1_+_79790705 | 0.24 |
ENSRNOT00000018233
|
Pglyrp1
|
peptidoglycan recognition protein 1 |
| chr7_-_99808612 | 0.23 |
ENSRNOT00000074953
|
AABR07058091.2
|
|
| chr1_+_254531659 | 0.23 |
ENSRNOT00000091377
|
AC080157.1
|
|
| chr19_+_24044103 | 0.19 |
ENSRNOT00000004621
|
Rnf150
|
ring finger protein 150 |
| chr7_-_82687130 | 0.18 |
ENSRNOT00000006791
|
Tmem74
|
transmembrane protein 74 |
| chr5_-_137104993 | 0.17 |
ENSRNOT00000027271
|
Ptprf
|
protein tyrosine phosphatase, receptor type, F |
| chr3_-_158328881 | 0.16 |
ENSRNOT00000044466
|
Ptprt
|
protein tyrosine phosphatase, receptor type, T |
| chr1_-_222350173 | 0.15 |
ENSRNOT00000030625
|
Flrt1
|
fibronectin leucine rich transmembrane protein 1 |
| chr7_+_25919867 | 0.15 |
ENSRNOT00000009625
ENSRNOT00000090153 |
Ric8b
|
RIC8 guanine nucleotide exchange factor B |
| chr5_+_14415841 | 0.15 |
ENSRNOT00000010682
|
Rgs20
|
regulator of G-protein signaling 20 |
| chr7_-_12393266 | 0.15 |
ENSRNOT00000021709
|
Efna2
|
ephrin A2 |
| chr9_-_30844199 | 0.15 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr5_+_27326762 | 0.15 |
ENSRNOT00000066191
|
Runx1t1
|
RUNX1 translocation partner 1 |
| chrX_+_39711201 | 0.14 |
ENSRNOT00000080512
ENSRNOT00000009802 |
Cnksr2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr13_+_51795867 | 0.14 |
ENSRNOT00000006747
|
Ube2t
|
ubiquitin-conjugating enzyme E2T |
| chr4_+_120672152 | 0.14 |
ENSRNOT00000077231
|
Mgll
|
monoglyceride lipase |
| chr3_+_15560712 | 0.14 |
ENSRNOT00000010218
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr15_+_61069581 | 0.14 |
ENSRNOT00000084333
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr6_-_107325345 | 0.13 |
ENSRNOT00000049481
ENSRNOT00000042594 ENSRNOT00000013026 |
Numb
|
NUMB, endocytic adaptor protein |
| chr1_-_215033460 | 0.13 |
ENSRNOT00000044565
|
Dusp8
|
dual specificity phosphatase 8 |
| chr7_+_64672722 | 0.13 |
ENSRNOT00000064448
ENSRNOT00000005539 |
Grip1
|
glutamate receptor interacting protein 1 |
| chr1_+_142679345 | 0.13 |
ENSRNOT00000034267
|
Zscan2
|
zinc finger and SCAN domain containing 2 |
| chr6_-_1319541 | 0.13 |
ENSRNOT00000006527
|
Strn
|
striatin |
| chr9_-_8632017 | 0.13 |
ENSRNOT00000043269
|
AABR07066416.1
|
|
| chr10_-_2037891 | 0.12 |
ENSRNOT00000004563
|
Ercc4
|
ERCC excision repair 4, endonuclease catalytic subunit |
| chr14_+_104250617 | 0.12 |
ENSRNOT00000079874
|
Spred2
|
sprouty-related, EVH1 domain containing 2 |
| chr1_-_71173216 | 0.12 |
ENSRNOT00000020523
|
Zfp28
|
zinc finger protein 28 |
| chr12_+_12624723 | 0.11 |
ENSRNOT00000092601
|
Ocm2
|
oncomodulin 2 |
| chr2_-_138833933 | 0.11 |
ENSRNOT00000013343
|
Pcdh18
|
protocadherin 18 |
| chr9_-_52238564 | 0.11 |
ENSRNOT00000005073
|
Col5a2
|
collagen type V alpha 2 chain |
| chr5_-_164927869 | 0.11 |
ENSRNOT00000012080
|
Draxin
|
dorsal inhibitory axon guidance protein |
| chr20_+_6054199 | 0.11 |
ENSRNOT00000092629
|
Brpf3
|
bromodomain and PHD finger containing, 3 |
| chr1_-_255171691 | 0.11 |
ENSRNOT00000090224
|
AC113773.2
|
|
| chr4_+_174692331 | 0.11 |
ENSRNOT00000011770
|
Plekha5
|
pleckstrin homology domain containing A5 |
| chr4_-_155740193 | 0.10 |
ENSRNOT00000043229
|
LOC102550396
|
LRRGT00188 |
| chr10_+_48903540 | 0.10 |
ENSRNOT00000004248
|
Trpv2
|
transient receptor potential cation channel, subfamily V, member 2 |
| chr13_+_34400170 | 0.10 |
ENSRNOT00000061516
ENSRNOT00000061515 ENSRNOT00000061513 ENSRNOT00000084506 ENSRNOT00000086641 |
Clasp1
|
cytoplasmic linker associated protein 1 |
| chr8_+_52729003 | 0.10 |
ENSRNOT00000081738
|
Nxpe4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr2_-_38110567 | 0.10 |
ENSRNOT00000072212
|
Ipo11
|
importin 11 |
| chr18_+_30487264 | 0.10 |
ENSRNOT00000040125
|
Pcdhb10
|
protocadherin beta 10 |
| chr15_-_43542939 | 0.10 |
ENSRNOT00000012996
|
Dpysl2
|
dihydropyrimidinase-like 2 |
| chr10_+_49259194 | 0.09 |
ENSRNOT00000091100
ENSRNOT00000004390 |
Fbxw10
|
F-box and WD repeat domain containing 10 |
| chr8_+_82037977 | 0.09 |
ENSRNOT00000082288
|
Myo5a
|
myosin VA |
| chr10_+_40543288 | 0.09 |
ENSRNOT00000016755
|
Slc36a1
|
solute carrier family 36 member 1 |
| chr18_+_30895831 | 0.09 |
ENSRNOT00000026985
|
Pcdhga10
|
protocadherin gamma subfamily A, 10 |
| chr3_-_38277440 | 0.09 |
ENSRNOT00000037857
|
Stam2
|
signal transducing adaptor molecule 2 |
| chr7_+_40217269 | 0.08 |
ENSRNOT00000082090
|
Cep290
|
centrosomal protein 290 |
| chr7_-_140502441 | 0.08 |
ENSRNOT00000089544
|
Kmt2d
|
lysine methyltransferase 2D |
| chr15_-_71779033 | 0.08 |
ENSRNOT00000017765
|
Pcdh20
|
protocadherin 20 |
| chr11_+_38035611 | 0.08 |
ENSRNOT00000087603
ENSRNOT00000002695 |
Mx2
|
MX dynamin like GTPase 2 |
| chr10_+_975697 | 0.08 |
ENSRNOT00000089404
|
AC117889.1
|
|
| chr2_+_116970344 | 0.08 |
ENSRNOT00000039603
|
Egfem1
|
EGF-like and EMI domain containing 1 |
| chr1_+_190462327 | 0.08 |
ENSRNOT00000030732
|
LOC691519
|
similar to ankyrin repeat domain 26 |
| chr18_+_30387937 | 0.08 |
ENSRNOT00000027210
|
Pcdhb4
|
protocadherin beta 4 |
| chr19_+_50848736 | 0.07 |
ENSRNOT00000077053
|
Cdh13
|
cadherin 13 |
| chr12_-_2592838 | 0.07 |
ENSRNOT00000079918
|
Evi5l
|
ecotropic viral integration site 5-like |
| chr1_+_144831523 | 0.07 |
ENSRNOT00000039748
|
Mex3b
|
mex-3 RNA binding family member B |
| chr13_-_67688477 | 0.07 |
ENSRNOT00000068148
|
Prg4
|
proteoglycan 4 |
| chr7_-_122403667 | 0.07 |
ENSRNOT00000088814
|
Mkl1
|
megakaryoblastic leukemia (translocation) 1 |
| chr3_-_143899370 | 0.07 |
ENSRNOT00000049635
|
Andpro
|
androgen regulated protein |
| chrX_-_119162518 | 0.07 |
ENSRNOT00000073176
|
LOC103694506
|
ubiquitin-conjugating enzyme E2 W |
| chr1_+_72882806 | 0.07 |
ENSRNOT00000024640
|
Tnni3
|
troponin I3, cardiac type |
| chr3_+_100786862 | 0.06 |
ENSRNOT00000080190
|
Bdnf
|
brain-derived neurotrophic factor |
| chr10_-_37215899 | 0.06 |
ENSRNOT00000006356
|
Sec24a
|
SEC24 homolog A, COPII coat complex component |
| chr8_+_82038967 | 0.06 |
ENSRNOT00000079535
|
Myo5a
|
myosin VA |
| chr20_+_48504264 | 0.06 |
ENSRNOT00000087740
|
Cdc40
|
cell division cycle 40 |
| chr6_-_125812517 | 0.06 |
ENSRNOT00000007061
|
Trip11
|
thyroid hormone receptor interactor 11 |
| chr8_-_21831668 | 0.06 |
ENSRNOT00000027897
|
Col5a3
|
collagen type V alpha 3 chain |
| chr1_+_88586837 | 0.06 |
ENSRNOT00000080555
|
Zfp382
|
zinc finger protein 382 |
| chr10_+_71536533 | 0.06 |
ENSRNOT00000088138
|
Acaca
|
acetyl-CoA carboxylase alpha |
| chr9_-_9142339 | 0.06 |
ENSRNOT00000046852
|
MGC116197
|
similar to RIKEN cDNA 1700001E04 |
| chr11_+_9642365 | 0.06 |
ENSRNOT00000087080
ENSRNOT00000042384 |
Robo1
|
roundabout guidance receptor 1 |
| chr3_-_93734282 | 0.06 |
ENSRNOT00000012428
|
Caprin1
|
cell cycle associated protein 1 |
| chr18_+_53088994 | 0.06 |
ENSRNOT00000091921
|
AC104053.2
|
|
| chr2_-_221099738 | 0.06 |
ENSRNOT00000022947
|
Snx7
|
sorting nexin 7 |
| chr10_+_83081168 | 0.06 |
ENSRNOT00000035023
|
Tac4
|
tachykinin 4 (hemokinin) |
| chr12_+_32103198 | 0.05 |
ENSRNOT00000085464
|
Tmem132d
|
transmembrane protein 132D |
| chr7_-_124367630 | 0.05 |
ENSRNOT00000055978
|
Ttll1
|
tubulin tyrosine ligase like 1 |
| chr4_-_28224858 | 0.05 |
ENSRNOT00000091075
|
AABR07059632.3
|
|
| chr15_+_39638510 | 0.05 |
ENSRNOT00000037800
|
Rcbtb1
|
RCC1 and BTB domain containing protein 1 |
| chr19_+_30794571 | 0.05 |
ENSRNOT00000024390
ENSRNOT00000083219 |
Gab1
|
GRB2-associated binding protein 1 |
| chr10_-_104748003 | 0.05 |
ENSRNOT00000042372
ENSRNOT00000046754 |
Acox1
|
acyl-CoA oxidase 1 |
| chr15_-_4399589 | 0.05 |
ENSRNOT00000065858
ENSRNOT00000079812 |
Fam149b1
|
family with sequence similarity 149, member B1 |
| chr2_+_85377318 | 0.05 |
ENSRNOT00000016506
ENSRNOT00000085094 |
Sema5a
|
semaphorin 5A |
| chr14_-_18862407 | 0.05 |
ENSRNOT00000003823
|
Cxcl6
|
C-X-C motif chemokine ligand 6 |
| chr6_+_86852323 | 0.05 |
ENSRNOT00000059244
|
Fancm
|
Fanconi anemia, complementation group M |
| chr5_+_106952082 | 0.05 |
ENSRNOT00000046931
|
RGD1561246
|
similar to put. precursor MuIFN-alpha 5 |
| chr2_-_196270826 | 0.05 |
ENSRNOT00000028609
|
Pip5k1a
|
phosphatidylinositol-4-phosphate 5-kinase, type 1, alpha |
| chr15_-_36798814 | 0.05 |
ENSRNOT00000065764
|
Cenpj
|
centromere protein J |
| chr1_-_214504569 | 0.04 |
ENSRNOT00000082221
|
Chid1
|
chitinase domain containing 1 |
| chr16_+_23668595 | 0.04 |
ENSRNOT00000067886
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr1_-_214067657 | 0.04 |
ENSRNOT00000044390
|
LOC100911881
|
chitinase domain-containing protein 1-like |
| chrX_+_156812064 | 0.04 |
ENSRNOT00000077142
|
Hcfc1
|
host cell factor C1 |
| chr11_-_87485833 | 0.04 |
ENSRNOT00000079948
|
AABR07034751.1
|
|
| chr2_-_34922721 | 0.04 |
ENSRNOT00000017832
|
Cwc27
|
CWC27 spliceosome-associated protein homolog |
| chr1_+_267618248 | 0.04 |
ENSRNOT00000017186
|
Gsto2
|
glutathione S-transferase omega 2 |
| chr18_+_57654290 | 0.03 |
ENSRNOT00000025892
|
Htr4
|
5-hydroxytryptamine receptor 4 |
| chr11_+_80319177 | 0.03 |
ENSRNOT00000036777
|
Rtp2
|
receptor (chemosensory) transporter protein 2 |
| chr8_-_21481735 | 0.03 |
ENSRNOT00000038069
|
Zfp426
|
zinc finger protein 426 |
| chr8_+_22368745 | 0.03 |
ENSRNOT00000049973
|
Slc44a2
|
solute carrier family 44 member 2 |
| chr1_-_233145924 | 0.03 |
ENSRNOT00000018763
ENSRNOT00000082442 |
Psat1
|
phosphoserine aminotransferase 1 |
| chr2_-_122690617 | 0.03 |
ENSRNOT00000018942
|
Mccc1
|
methylcrotonoyl-CoA carboxylase 1 |
| chr1_+_72356880 | 0.03 |
ENSRNOT00000021917
|
Zfp865
|
zinc finger protein 865 |
| chr15_+_27739251 | 0.03 |
ENSRNOT00000011840
|
Parp2
|
poly (ADP-ribose) polymerase 2 |
| chr13_-_74520634 | 0.03 |
ENSRNOT00000077169
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr10_+_86711240 | 0.03 |
ENSRNOT00000012812
|
Msl1
|
male specific lethal 1 homolog |
| chr1_+_91152635 | 0.03 |
ENSRNOT00000073438
|
LOC100912070
|
dermokine-like |
| chr1_+_101397828 | 0.03 |
ENSRNOT00000028189
|
Kcna7
|
potassium voltage-gated channel subfamily A member 7 |
| chr15_-_28155291 | 0.02 |
ENSRNOT00000050820
|
Rnase2
|
ribonuclease A family member 2 |
| chr14_-_37361798 | 0.02 |
ENSRNOT00000002980
|
Cwh43
|
cell wall biogenesis 43 C-terminal homolog |
| chr1_+_174655939 | 0.02 |
ENSRNOT00000014002
|
Ipo7
|
importin 7 |
| chr5_+_172259520 | 0.02 |
ENSRNOT00000037211
|
Ttc34
|
tetratricopeptide repeat domain 34 |
| chr2_-_149563905 | 0.02 |
ENSRNOT00000086186
|
Igsf10
|
immunoglobulin superfamily, member 10 |
| chr6_-_23581052 | 0.02 |
ENSRNOT00000006190
|
Ppp1cb
|
protein phosphatase 1 catalytic subunit beta |
| chr1_+_169590308 | 0.02 |
ENSRNOT00000023147
|
Olr155
|
olfactory receptor 155 |
| chrX_-_159841072 | 0.02 |
ENSRNOT00000001158
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor 6 |
| chr12_+_13508933 | 0.02 |
ENSRNOT00000083976
|
Rnf216
|
ring finger protein 216 |
| chr10_-_45514878 | 0.02 |
ENSRNOT00000036940
|
Iba57
|
IBA57 homolog, iron-sulfur cluster assembly |
| chr1_-_171979933 | 0.02 |
ENSRNOT00000026744
|
Cyb5r2
|
cytochrome b5 reductase 2 |
| chr1_-_264294630 | 0.02 |
ENSRNOT00000035758
|
Sec31b
|
SEC31 homolog B, COPII coat complex component |
| chr12_+_8032809 | 0.02 |
ENSRNOT00000082850
|
Slc7a1
|
solute carrier family 7 member 1 |
| chr2_-_22105710 | 0.02 |
ENSRNOT00000017705
|
Zfyve16
|
zinc finger FYVE-type containing 16 |
| chr10_-_107324355 | 0.02 |
ENSRNOT00000064450
|
Usp36
|
ubiquitin specific peptidase 36 |
| chr4_+_170820594 | 0.02 |
ENSRNOT00000032568
|
LOC500354
|
similar to C030030A07Rik protein |
| chr8_+_85259982 | 0.02 |
ENSRNOT00000010409
|
Elovl5
|
ELOVL fatty acid elongase 5 |
| chr7_+_70335061 | 0.02 |
ENSRNOT00000072176
ENSRNOT00000084933 |
Cyp27b1
|
cytochrome P450, family 27, subfamily b, polypeptide 1 |
| chr18_+_30808404 | 0.02 |
ENSRNOT00000060475
|
Pcdhga1
|
protocadherin gamma subfamily A, 1 |
| chr7_+_12433933 | 0.02 |
ENSRNOT00000060690
|
Cbarp
|
CACN beta subunit associated regulatory protein |
| chr8_+_79638696 | 0.02 |
ENSRNOT00000085959
|
Dyx1c1
|
dyslexia susceptibility 1 candidate 1 |
| chr6_-_26241337 | 0.01 |
ENSRNOT00000006525
|
Slc4a1ap
|
solute carrier family 4 member 1 adaptor protein |
| chr7_+_80625010 | 0.01 |
ENSRNOT00000084940
|
Oxr1
|
oxidation resistance 1 |
| chr10_-_70681672 | 0.01 |
ENSRNOT00000013836
|
Mmp28
|
matrix metallopeptidase 28 |
| chr15_-_24374753 | 0.01 |
ENSRNOT00000016274
|
Atg14
|
autophagy related 14 |
| chr16_-_18645941 | 0.01 |
ENSRNOT00000079241
|
Dydc2
|
DPY30 domain containing 2 |
| chrM_-_14061 | 0.01 |
ENSRNOT00000051268
|
Mt-nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr2_+_18937458 | 0.01 |
ENSRNOT00000022377
|
Tmem167a
|
transmembrane protein 167A |
| chr2_+_27003783 | 0.01 |
ENSRNOT00000061507
|
Poc5
|
POC5 centriolar protein |
| chr3_-_45210474 | 0.01 |
ENSRNOT00000091777
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr2_-_45480798 | 0.01 |
ENSRNOT00000066764
|
Snx18
|
sorting nexin 18 |
| chr20_+_48503973 | 0.01 |
ENSRNOT00000064081
|
Cdc40
|
cell division cycle 40 |
| chr3_+_152294656 | 0.01 |
ENSRNOT00000027067
|
Phf20
|
PHD finger protein 20 |
| chr15_+_28439317 | 0.01 |
ENSRNOT00000060519
|
LOC690384
|
similar to ribosomal protein L31 |
| chr10_-_88036040 | 0.01 |
ENSRNOT00000018851
|
Krt13
|
keratin 13 |
| chr2_-_149564113 | 0.01 |
ENSRNOT00000018628
|
Igsf10
|
immunoglobulin superfamily, member 10 |
| chrX_+_31054394 | 0.01 |
ENSRNOT00000037439
|
LOC688526
|
similar to ribonucleic acid binding protein S1 |
| chr18_-_30781292 | 0.01 |
ENSRNOT00000027125
|
Taf7
|
TATA-box binding protein associated factor 7 |
| chr1_-_171979757 | 0.01 |
ENSRNOT00000079844
|
Cyb5r2
|
cytochrome b5 reductase 2 |
| chr9_-_17880706 | 0.01 |
ENSRNOT00000031549
|
Aars2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr9_-_92775816 | 0.01 |
ENSRNOT00000029635
|
RGD1563917
|
similar to Nuclear autoantigen Sp-100 (Speckled 100 kDa) |
| chr1_-_88723973 | 0.00 |
ENSRNOT00000082555
|
NEWGENE_1306714
|
WD repeat domain 62-like 1 |
| chr19_-_43528851 | 0.00 |
ENSRNOT00000036972
|
Mlkl
|
mixed lineage kinase domain like pseudokinase |
| chr4_+_98457810 | 0.00 |
ENSRNOT00000074175
|
AABR07060872.1
|
|
| chr19_+_57286985 | 0.00 |
ENSRNOT00000058074
|
Cog2
|
component of oligomeric golgi complex 2 |
| chr12_+_11240761 | 0.00 |
ENSRNOT00000001310
|
Pdap1
|
PDGFA associated protein 1 |
| chr9_-_55256340 | 0.00 |
ENSRNOT00000028907
|
Sdpr
|
serum deprivation response |
| chr19_+_55206455 | 0.00 |
ENSRNOT00000039273
|
Zc3h18
|
zinc finger CCCH-type containing 18 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ddit3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032197 | transposition, RNA-mediated(GO:0032197) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.1 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.2 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.2 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.1 | GO:0048691 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.0 | 0.2 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 | 0.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.1 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.0 | GO:0070949 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.0 | 0.1 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.1 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.0 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.1 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.0 | GO:0019046 | viral latency(GO:0019042) release from viral latency(GO:0019046) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.0 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |