Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
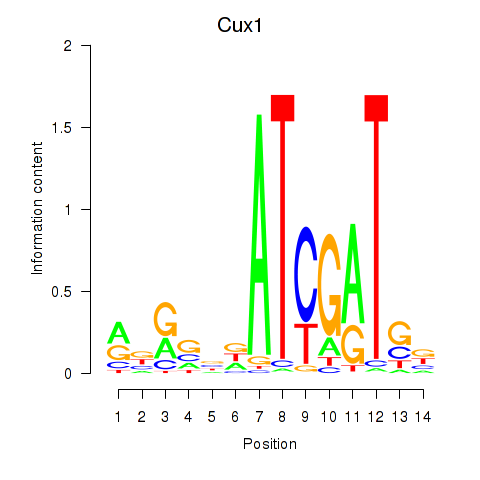
Results for Cux1
Z-value: 0.73
Transcription factors associated with Cux1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cux1
|
ENSRNOG00000001424 | cut-like homeobox 1 |
|
Cux1
|
ENSRNOG00000059116 | cut-like homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cux1 | rn6_v1_chr12_+_23151180_23151180 | -0.01 | 9.8e-01 | Click! |
Activity profile of Cux1 motif
Sorted Z-values of Cux1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_22859622 | 0.37 |
ENSRNOT00000073501
ENSRNOT00000068410 |
Adam22
|
ADAM metallopeptidase domain 22 |
| chr19_-_39267928 | 0.34 |
ENSRNOT00000027686
|
Tmed6
|
transmembrane p24 trafficking protein 6 |
| chr2_-_157058194 | 0.33 |
ENSRNOT00000076409
|
Rn50_2_1765.1
|
|
| chr15_-_30147793 | 0.31 |
ENSRNOT00000060399
|
AABR07017693.1
|
|
| chr15_+_36865548 | 0.30 |
ENSRNOT00000076460
|
Parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chr5_+_142875773 | 0.27 |
ENSRNOT00000082120
ENSRNOT00000056496 |
Epha10
|
EPH receptor A10 |
| chr11_-_60882379 | 0.27 |
ENSRNOT00000002799
|
Cd200r1
|
CD200 receptor 1 |
| chr2_+_23289374 | 0.26 |
ENSRNOT00000090666
ENSRNOT00000032783 |
Dmgdh
|
dimethylglycine dehydrogenase |
| chr11_-_62451149 | 0.26 |
ENSRNOT00000093686
ENSRNOT00000081443 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr4_+_61814974 | 0.23 |
ENSRNOT00000074951
|
Akr1b10
|
aldo-keto reductase family 1 member B10 |
| chr2_-_220838905 | 0.23 |
ENSRNOT00000078914
|
AABR07013065.1
|
|
| chr6_+_64224861 | 0.23 |
ENSRNOT00000093159
ENSRNOT00000093664 |
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr8_-_22150005 | 0.21 |
ENSRNOT00000041678
|
Tyk2
|
tyrosine kinase 2 |
| chrM_+_11736 | 0.21 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr3_+_114355798 | 0.21 |
ENSRNOT00000024658
ENSRNOT00000036435 |
Slc28a2
|
solute carrier family 28 member 2 |
| chr10_+_31647898 | 0.20 |
ENSRNOT00000066536
|
LOC689968
|
similar to kidney injury molecule 1 |
| chr1_+_213583606 | 0.20 |
ENSRNOT00000088899
|
AC109844.1
|
|
| chr13_+_51534025 | 0.20 |
ENSRNOT00000006637
|
Syt2
|
synaptotagmin 2 |
| chrX_-_4945944 | 0.19 |
ENSRNOT00000077238
|
Kdm6a
|
lysine demethylase 6A |
| chr4_+_152450126 | 0.19 |
ENSRNOT00000076930
|
Rad52
|
RAD52 homolog, DNA repair protein |
| chr2_-_192381716 | 0.19 |
ENSRNOT00000064950
|
AABR07012291.1
|
|
| chr13_-_47397890 | 0.19 |
ENSRNOT00000005505
|
C4bpb
|
complement component 4 binding protein, beta |
| chr2_+_86891092 | 0.17 |
ENSRNOT00000077162
ENSRNOT00000083066 |
LOC100360380
|
zinc finger protein 457-like |
| chr2_-_257546799 | 0.17 |
ENSRNOT00000089370
ENSRNOT00000090367 |
Miga1
|
mitoguardin 1 |
| chr6_+_99356509 | 0.16 |
ENSRNOT00000008416
|
Akap5
|
A-kinase anchoring protein 5 |
| chr8_-_69466618 | 0.16 |
ENSRNOT00000042925
|
Ids
|
iduronate 2-sulfatase |
| chr7_+_20262680 | 0.16 |
ENSRNOT00000046378
|
LOC300308
|
similar to hypothetical protein 4930509O22 |
| chr20_-_7654428 | 0.15 |
ENSRNOT00000045044
|
Tcp11
|
t-complex 11 |
| chr8_+_12355767 | 0.15 |
ENSRNOT00000068445
|
Fam76b
|
family with sequence similarity 76, member B |
| chr15_+_47470863 | 0.15 |
ENSRNOT00000072438
|
LOC683422
|
similar to Plasma kallikrein precursor (Plasma prekallikrein) (Kininogenin) (Fletcher factor) |
| chr8_+_48438259 | 0.15 |
ENSRNOT00000059813
|
Mfrp
|
membrane frizzled-related protein |
| chr10_-_85684138 | 0.15 |
ENSRNOT00000017989
|
Pip4k2b
|
phosphatidylinositol-5-phosphate 4-kinase type 2 beta |
| chr17_-_69711689 | 0.15 |
ENSRNOT00000041925
|
Akr1c12
|
aldo-keto reductase family 1, member C12 |
| chrX_-_70460536 | 0.14 |
ENSRNOT00000076824
|
Pdzd11
|
PDZ domain containing 11 |
| chr8_-_5429581 | 0.14 |
ENSRNOT00000044174
|
Dync2h1
|
dynein cytoplasmic 2 heavy chain 1 |
| chr17_+_16455026 | 0.14 |
ENSRNOT00000022932
|
Zfp169
|
zinc finger protein 169 |
| chr9_+_66889028 | 0.14 |
ENSRNOT00000087194
|
Carf
|
calcium responsive transcription factor |
| chr10_+_66942398 | 0.14 |
ENSRNOT00000018986
|
Rab11fip4
|
RAB11 family interacting protein 4 |
| chr9_+_49903085 | 0.14 |
ENSRNOT00000022601
|
RGD1310553
|
similar to expressed sequence AI597479 |
| chrX_+_137934484 | 0.13 |
ENSRNOT00000049046
|
LOC317588
|
hypothetical protein LOC317588 |
| chr16_-_19097329 | 0.13 |
ENSRNOT00000030617
|
RGD1311847
|
similar to 1700030K09Rik protein |
| chr1_-_43638161 | 0.13 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr14_-_67170361 | 0.13 |
ENSRNOT00000005477
|
Slit2
|
slit guidance ligand 2 |
| chrX_-_78911601 | 0.13 |
ENSRNOT00000003188
|
RGD1566265
|
similar to RIKEN cDNA 2610002M06 |
| chr3_-_119330345 | 0.13 |
ENSRNOT00000077398
ENSRNOT00000079191 |
Trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr3_-_146396299 | 0.12 |
ENSRNOT00000040188
ENSRNOT00000008931 |
Apmap
|
adipocyte plasma membrane associated protein |
| chr3_+_113918629 | 0.12 |
ENSRNOT00000078978
ENSRNOT00000037168 |
Ctdspl2
|
CTD small phosphatase like 2 |
| chr4_+_2053712 | 0.12 |
ENSRNOT00000045086
|
Rnf32
|
ring finger protein 32 |
| chr7_+_127081978 | 0.12 |
ENSRNOT00000022945
|
Tbc1d22a
|
TBC1 domain family, member 22a |
| chr19_+_14489983 | 0.12 |
ENSRNOT00000075435
|
Zfp14
|
ZFP14 zinc finger protein |
| chr18_-_56500941 | 0.12 |
ENSRNOT00000024704
|
Hmgxb3
|
HMG-box containing 3 |
| chr6_-_115616766 | 0.12 |
ENSRNOT00000006143
ENSRNOT00000045870 |
Sel1l
|
SEL1L ERAD E3 ligase adaptor subunit |
| chr5_-_137125737 | 0.12 |
ENSRNOT00000088532
|
AABR07049768.1
|
|
| chr9_-_83253458 | 0.12 |
ENSRNOT00000041689
|
Epha4
|
Eph receptor A4 |
| chr11_-_31180616 | 0.11 |
ENSRNOT00000065535
ENSRNOT00000052033 ENSRNOT00000002812 |
Synj1
|
synaptojanin 1 |
| chr8_-_96547568 | 0.11 |
ENSRNOT00000078343
|
RGD1560775
|
similar to RIKEN cDNA 4930579C12 gene |
| chr9_-_92530938 | 0.11 |
ENSRNOT00000064875
|
Slc16a14
|
solute carrier family 16, member 14 |
| chr10_-_46720907 | 0.11 |
ENSRNOT00000083093
ENSRNOT00000067866 |
Tom1l2
|
target of myb1 like 2 membrane trafficking protein |
| chr1_-_81043340 | 0.11 |
ENSRNOT00000051473
|
Zfp111
|
zinc finger protein 111 |
| chr3_+_147073160 | 0.11 |
ENSRNOT00000012690
|
Sdcbp2
|
syndecan binding protein 2 |
| chr14_+_39368530 | 0.11 |
ENSRNOT00000084367
|
Cox7b2
|
cytochrome c oxidase subunit 7B2 |
| chr4_+_179481263 | 0.11 |
ENSRNOT00000021284
|
Etfrf1
|
electron transfer flavoprotein regulatory factor 1 |
| chr12_-_18120476 | 0.11 |
ENSRNOT00000080364
|
RGD1566386
|
similar to Hypothetical protein A430033K04 |
| chr12_+_8725517 | 0.11 |
ENSRNOT00000001243
|
Slc46a3
|
solute carrier family 46, member 3 |
| chr7_-_135800481 | 0.11 |
ENSRNOT00000029097
|
Pus7l
|
pseudouridylate synthase 7-like |
| chr12_+_9360672 | 0.11 |
ENSRNOT00000088957
|
Flt3
|
fms-related tyrosine kinase 3 |
| chr7_-_94774569 | 0.11 |
ENSRNOT00000036399
|
Dscc1
|
DNA replication and sister chromatid cohesion 1 |
| chr3_-_55951584 | 0.11 |
ENSRNOT00000036585
|
Fastkd1
|
FAST kinase domains 1 |
| chr10_+_83081168 | 0.10 |
ENSRNOT00000035023
|
Tac4
|
tachykinin 4 (hemokinin) |
| chrX_+_28072826 | 0.10 |
ENSRNOT00000039796
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr3_+_79823945 | 0.10 |
ENSRNOT00000014484
|
Celf1
|
CUGBP, Elav-like family member 1 |
| chr12_-_5773036 | 0.10 |
ENSRNOT00000041365
|
Fry
|
FRY microtubule binding protein |
| chr3_+_63379031 | 0.10 |
ENSRNOT00000068199
|
Osbpl6
|
oxysterol binding protein-like 6 |
| chr10_-_37215899 | 0.10 |
ENSRNOT00000006356
|
Sec24a
|
SEC24 homolog A, COPII coat complex component |
| chr17_-_86657473 | 0.10 |
ENSRNOT00000078827
|
AABR07028795.1
|
|
| chr2_-_236480502 | 0.10 |
ENSRNOT00000015020
|
Sgms2
|
sphingomyelin synthase 2 |
| chr8_+_48437918 | 0.10 |
ENSRNOT00000085578
|
Mfrp
|
membrane frizzled-related protein |
| chr17_+_35396286 | 0.10 |
ENSRNOT00000089613
ENSRNOT00000092638 |
Exoc2
|
exocyst complex component 2 |
| chr11_+_67555658 | 0.10 |
ENSRNOT00000039075
|
Csta
|
cystatin A (stefin A) |
| chr6_+_86823684 | 0.10 |
ENSRNOT00000086081
ENSRNOT00000006574 |
Fancm
|
Fanconi anemia, complementation group M |
| chr16_+_11599753 | 0.10 |
ENSRNOT00000079833
|
Grid1
|
glutamate ionotropic receptor delta type subunit 1 |
| chr1_+_278557792 | 0.10 |
ENSRNOT00000023414
|
Atrnl1
|
attractin like 1 |
| chr3_-_9936352 | 0.10 |
ENSRNOT00000011224
ENSRNOT00000042798 |
Fnbp1
|
formin binding protein 1 |
| chr12_-_35979193 | 0.10 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr8_-_36467627 | 0.10 |
ENSRNOT00000082346
|
Fam118b
|
family with sequence similarity 118, member B |
| chr6_-_108796124 | 0.10 |
ENSRNOT00000086545
|
Arel1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr11_+_38727048 | 0.10 |
ENSRNOT00000081537
|
LOC103690343
|
zinc finger protein 260-like |
| chr9_-_100479868 | 0.10 |
ENSRNOT00000022748
|
Pask
|
PAS domain containing serine/threonine kinase |
| chr2_+_34312766 | 0.10 |
ENSRNOT00000060962
|
Cenpk
|
centromere protein K |
| chr8_+_32452885 | 0.10 |
ENSRNOT00000010423
|
Prdm10
|
PR/SET domain 10 |
| chr9_-_89195273 | 0.10 |
ENSRNOT00000022281
|
Sphkap
|
SPHK1 interactor, AKAP domain containing |
| chr3_+_79498179 | 0.10 |
ENSRNOT00000030750
|
Nup160
|
nucleoporin 160 |
| chr2_-_54963448 | 0.10 |
ENSRNOT00000017886
|
Ptger4
|
prostaglandin E receptor 4 |
| chr15_-_58171049 | 0.10 |
ENSRNOT00000001371
|
Gpalpp1
|
GPALPP motifs containing 1 |
| chr10_-_87153982 | 0.10 |
ENSRNOT00000014414
|
Krt222
|
keratin 222 |
| chr7_+_125288081 | 0.10 |
ENSRNOT00000085216
|
Parvg
|
parvin, gamma |
| chr18_-_6587080 | 0.09 |
ENSRNOT00000040815
|
LOC103694404
|
60S ribosomal protein L39 |
| chr1_-_263885169 | 0.09 |
ENSRNOT00000030782
|
Chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr8_+_116771982 | 0.09 |
ENSRNOT00000083240
|
AC128059.3
|
|
| chr20_-_26589209 | 0.09 |
ENSRNOT00000049437
|
Ctnna3
|
catenin alpha 3 |
| chr1_+_196839321 | 0.09 |
ENSRNOT00000020388
|
Kdm8
|
lysine demethylase 8 |
| chr5_+_43603043 | 0.09 |
ENSRNOT00000009899
|
Epha7
|
Eph receptor A7 |
| chr16_-_81756654 | 0.09 |
ENSRNOT00000026653
|
Cul4a
|
cullin 4A |
| chr7_+_15785410 | 0.09 |
ENSRNOT00000082664
ENSRNOT00000073235 |
Zfp955a
|
zinc finger protein 955A |
| chr18_+_30900291 | 0.09 |
ENSRNOT00000060461
|
Pcdhgb7
|
protocadherin gamma subfamily B, 7 |
| chr7_-_2961873 | 0.09 |
ENSRNOT00000067441
|
Rps15-ps2
|
ribosomal protein S15, pseudogene 2 |
| chr3_-_66279155 | 0.09 |
ENSRNOT00000079887
|
Cerkl
|
ceramide kinase-like |
| chr7_-_15073052 | 0.09 |
ENSRNOT00000037708
|
Zfp799
|
zinc finger protein 799 |
| chr5_+_5616483 | 0.09 |
ENSRNOT00000011026
|
Ncoa2
|
nuclear receptor coactivator 2 |
| chr7_+_27174882 | 0.09 |
ENSRNOT00000051181
|
Glt8d2
|
glycosyltransferase 8 domain containing 2 |
| chr2_+_118547190 | 0.09 |
ENSRNOT00000083676
ENSRNOT00000090301 ENSRNOT00000013410 |
Kcnmb2
|
potassium calcium-activated channel subfamily M regulatory beta subunit 2 |
| chr10_-_38969501 | 0.09 |
ENSRNOT00000090691
ENSRNOT00000081309 ENSRNOT00000010029 |
Il4
|
interleukin 4 |
| chr1_-_37741709 | 0.09 |
ENSRNOT00000079186
|
Fastkd3
|
FAST kinase domains 3 |
| chr13_-_48400632 | 0.09 |
ENSRNOT00000074204
|
Avpr1b
|
arginine vasopressin receptor 1B |
| chr14_+_100373129 | 0.09 |
ENSRNOT00000029980
|
Wdr92
|
WD repeat domain 92 |
| chr3_-_90751055 | 0.09 |
ENSRNOT00000040741
|
LOC499843
|
LRRGT00091 |
| chr17_-_9558624 | 0.09 |
ENSRNOT00000036223
ENSRNOT00000083492 |
B4galt7
|
beta-1,4-galactosyltransferase 7 |
| chr2_-_170460754 | 0.09 |
ENSRNOT00000013009
|
Slitrk3
|
SLIT and NTRK-like family, member 3 |
| chrX_-_14331486 | 0.09 |
ENSRNOT00000067603
|
Rpgr
|
retinitis pigmentosa GTPase regulator |
| chr1_+_228684136 | 0.08 |
ENSRNOT00000028608
|
Olr337
|
olfactory receptor 337 |
| chr8_-_71728654 | 0.08 |
ENSRNOT00000031207
ENSRNOT00000091751 |
Snx22
|
sorting nexin 22 |
| chr9_+_73528681 | 0.08 |
ENSRNOT00000084340
|
Unc80
|
unc-80 homolog, NALCN activator |
| chr1_+_98228573 | 0.08 |
ENSRNOT00000019278
|
LOC102555672
|
zinc finger protein 850-like |
| chr4_+_98648545 | 0.08 |
ENSRNOT00000008451
|
Eif2ak3
|
eukaryotic translation initiation factor 2 alpha kinase 3 |
| chr8_+_52729003 | 0.08 |
ENSRNOT00000081738
|
Nxpe4
|
neurexophilin and PC-esterase domain family, member 4 |
| chrX_-_37705263 | 0.08 |
ENSRNOT00000043666
|
Map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr13_+_97838361 | 0.08 |
ENSRNOT00000003641
|
Cnst
|
consortin, connexin sorting protein |
| chr14_-_8600512 | 0.08 |
ENSRNOT00000092537
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr17_+_9736577 | 0.08 |
ENSRNOT00000066586
|
F12
|
coagulation factor XII |
| chr19_+_37873515 | 0.08 |
ENSRNOT00000026163
ENSRNOT00000089921 |
Pskh1
|
protein serine kinase H1 |
| chr20_-_27301952 | 0.08 |
ENSRNOT00000000438
|
Slc25a16
|
solute carrier family 25 member 16 |
| chr20_+_9791171 | 0.08 |
ENSRNOT00000078031
|
Abcg1
|
ATP binding cassette subfamily G member 1 |
| chr2_-_219628997 | 0.08 |
ENSRNOT00000064484
|
Trmt13
|
tRNA methyltransferase 13 homolog |
| chr10_-_49149479 | 0.08 |
ENSRNOT00000004291
|
Zfp287
|
zinc finger protein 287 |
| chr3_+_37599728 | 0.08 |
ENSRNOT00000090575
|
Rif1
|
replication timing regulatory factor 1 |
| chr2_-_115891097 | 0.08 |
ENSRNOT00000013191
|
Skil
|
SKI-like proto-oncogene |
| chr2_-_27287605 | 0.08 |
ENSRNOT00000034041
|
Ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chr4_+_114918488 | 0.08 |
ENSRNOT00000014249
|
Slc4a5
|
solute carrier family 4 member 5 |
| chrX_-_105308554 | 0.08 |
ENSRNOT00000045556
|
Taf7l
|
TATA-box binding protein associated factor 7-like |
| chr8_+_19888667 | 0.08 |
ENSRNOT00000078593
|
Zfp317
|
zinc finger protein 317 |
| chr1_-_82344345 | 0.08 |
ENSRNOT00000051892
ENSRNOT00000090629 ENSRNOT00000029487 ENSRNOT00000027933 |
Ceacam1
|
carcinoembryonic antigen related cell adhesion molecule 1 |
| chr15_+_44411865 | 0.08 |
ENSRNOT00000017566
|
Kctd9
|
potassium channel tetramerization domain containing 9 |
| chr15_-_57805184 | 0.08 |
ENSRNOT00000000168
|
Cog3
|
component of oligomeric golgi complex 3 |
| chr14_+_71542057 | 0.08 |
ENSRNOT00000082592
ENSRNOT00000083701 ENSRNOT00000084322 |
Prom1
|
prominin 1 |
| chr3_+_155160481 | 0.08 |
ENSRNOT00000021133
|
Ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr15_+_87722221 | 0.08 |
ENSRNOT00000082688
|
Scel
|
sciellin |
| chr9_-_20154077 | 0.08 |
ENSRNOT00000082904
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr7_-_15072703 | 0.08 |
ENSRNOT00000087294
|
Zfp799
|
zinc finger protein 799 |
| chr8_+_69127708 | 0.08 |
ENSRNOT00000013490
|
Snapc5
|
small nuclear RNA activating complex, polypeptide 5 |
| chr7_-_139835876 | 0.08 |
ENSRNOT00000014258
|
Olr1877
|
olfactory receptor 1877 |
| chr16_-_7588841 | 0.07 |
ENSRNOT00000084645
|
Mettl6
|
methyltransferase like 6 |
| chr4_+_68656928 | 0.07 |
ENSRNOT00000016232
|
Tas2r108
|
taste receptor, type 2, member 108 |
| chr3_-_2770620 | 0.07 |
ENSRNOT00000008253
|
Traf2
|
Tnf receptor-associated factor 2 |
| chr10_+_11090314 | 0.07 |
ENSRNOT00000086305
|
Coro7
|
coronin 7 |
| chr14_-_46529375 | 0.07 |
ENSRNOT00000077306
ENSRNOT00000078682 |
LOC257642
|
rRNA promoter binding protein |
| chr20_-_13994794 | 0.07 |
ENSRNOT00000093466
|
Ggt5
|
gamma-glutamyltransferase 5 |
| chr10_-_72235888 | 0.07 |
ENSRNOT00000039481
|
Znhit3
|
zinc finger, HIT-type containing 3 |
| chr11_+_67101714 | 0.07 |
ENSRNOT00000078936
|
Cd86
|
CD86 molecule |
| chr20_-_8202924 | 0.07 |
ENSRNOT00000071399
|
Tmem217
|
transmembrane protein 217 |
| chr12_-_48254822 | 0.07 |
ENSRNOT00000056865
|
Ung
|
uracil-DNA glycosylase |
| chr14_+_32257805 | 0.07 |
ENSRNOT00000039626
|
AABR07014804.1
|
|
| chr10_+_86223336 | 0.07 |
ENSRNOT00000038939
|
LOC257650
|
hippyragranin |
| chr7_+_123043503 | 0.07 |
ENSRNOT00000026258
ENSRNOT00000086355 |
Tef
|
TEF, PAR bZIP transcription factor |
| chr7_+_130474279 | 0.07 |
ENSRNOT00000092388
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr1_-_65611815 | 0.07 |
ENSRNOT00000036874
|
Zfp324
|
zinc finger protein 324 |
| chr17_-_66295014 | 0.07 |
ENSRNOT00000023974
|
Mtr
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr10_-_36322356 | 0.07 |
ENSRNOT00000000248
|
Zfp354c
|
zinc finger protein 354C |
| chr11_+_83855753 | 0.07 |
ENSRNOT00000030686
|
RGD1563956
|
similar to 60S ribosomal protein L12 |
| chr3_-_16537433 | 0.07 |
ENSRNOT00000048523
|
AABR07051533.2
|
|
| chr18_-_56728185 | 0.07 |
ENSRNOT00000066048
|
Ppargc1b
|
PPARG coactivator 1 beta |
| chr9_-_5330815 | 0.07 |
ENSRNOT00000014548
|
Slc5a7
|
solute carrier family 5 member 7 |
| chr8_-_12355091 | 0.07 |
ENSRNOT00000009318
|
Cep57
|
centrosomal protein 57 |
| chr9_+_10603813 | 0.07 |
ENSRNOT00000073991
ENSRNOT00000074469 ENSRNOT00000079999 |
Ptprs
|
protein tyrosine phosphatase, receptor type, S |
| chr1_+_128614138 | 0.07 |
ENSRNOT00000076227
ENSRNOT00000078707 |
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr5_-_140657745 | 0.07 |
ENSRNOT00000019080
|
Mfsd2a
|
major facilitator superfamily domain containing 2A |
| chr8_+_85259982 | 0.07 |
ENSRNOT00000010409
|
Elovl5
|
ELOVL fatty acid elongase 5 |
| chr1_-_263910251 | 0.07 |
ENSRNOT00000017202
|
Cwf19l1
|
CWF19-like 1, cell cycle control (S. pombe) |
| chr5_-_169630340 | 0.07 |
ENSRNOT00000087043
|
Kcnab2
|
potassium voltage-gated channel subfamily A regulatory beta subunit 2 |
| chr18_-_75207306 | 0.07 |
ENSRNOT00000021717
|
Setbp1
|
SET binding protein 1 |
| chr5_-_117612123 | 0.07 |
ENSRNOT00000065112
|
Dock7
|
dedicator of cytokinesis 7 |
| chr11_-_66878551 | 0.07 |
ENSRNOT00000066616
|
Iqcb1
|
IQ motif containing B1 |
| chr13_-_26812215 | 0.07 |
ENSRNOT00000003728
|
Kdsr
|
3-ketodihydrosphingosine reductase |
| chr1_+_116604550 | 0.07 |
ENSRNOT00000083587
|
Ube3a
|
ubiquitin protein ligase E3A |
| chr8_+_58120179 | 0.07 |
ENSRNOT00000049076
ENSRNOT00000090114 |
Npat
|
nuclear protein, co-activator of histone transcription |
| chr19_+_755460 | 0.07 |
ENSRNOT00000076560
|
Dync1li2
|
dynein, cytoplasmic 1 light intermediate chain 2 |
| chr14_-_46657975 | 0.07 |
ENSRNOT00000087247
|
LOC257642
|
rRNA promoter binding protein |
| chr20_+_12944786 | 0.07 |
ENSRNOT00000048218
|
Pcnt
|
pericentrin |
| chr4_-_6062641 | 0.07 |
ENSRNOT00000074846
|
Cct8l1
|
chaperonin containing TCP1, subunit 8 (theta)-like 1 |
| chr10_-_91661558 | 0.07 |
ENSRNOT00000043156
|
AABR07030521.1
|
|
| chr2_-_188718704 | 0.06 |
ENSRNOT00000028010
|
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr5_-_138545404 | 0.06 |
ENSRNOT00000011586
|
Rimkla
|
ribosomal modification protein rimK-like family member A |
| chr18_+_57516347 | 0.06 |
ENSRNOT00000082215
|
Gm9949
|
predicted gene 9949 |
| chr5_-_78985990 | 0.06 |
ENSRNOT00000009248
|
Ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr13_+_90301006 | 0.06 |
ENSRNOT00000029315
|
Slamf6
|
SLAM family member 6 |
| chr4_+_60549197 | 0.06 |
ENSRNOT00000071249
ENSRNOT00000075621 |
Exoc4
|
exocyst complex component 4 |
| chr20_+_20105047 | 0.06 |
ENSRNOT00000082181
|
Ank3
|
ankyrin 3 |
| chr5_+_36555109 | 0.06 |
ENSRNOT00000061128
|
Fbxl4
|
F-box and leucine-rich repeat protein 4 |
| chr5_-_50068706 | 0.06 |
ENSRNOT00000084643
|
Orc3
|
origin recognition complex, subunit 3 |
| chr7_+_71293388 | 0.06 |
ENSRNOT00000083959
|
Ptdss1
|
phosphatidylserine synthase 1 |
| chr15_+_3754262 | 0.06 |
ENSRNOT00000035100
|
NEWGENE_1304700
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Cux1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.1 | 0.2 | GO:0045959 | negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.1 | 0.2 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) polyprenol catabolic process(GO:0016095) |
| 0.1 | 0.1 | GO:0072703 | response to methyl methanesulfonate(GO:0072702) cellular response to methyl methanesulfonate(GO:0072703) |
| 0.1 | 0.2 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 0.2 | GO:0036394 | amylase secretion(GO:0036394) |
| 0.1 | 0.2 | GO:0072642 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.0 | 0.1 | GO:0032197 | transposition, RNA-mediated(GO:0032197) |
| 0.0 | 0.3 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.0 | 0.1 | GO:0071623 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 0.0 | 0.3 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.0 | 0.1 | GO:0002353 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:2001106 | fasciculation of motor neuron axon(GO:0097156) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 | 0.1 | GO:0043316 | cytotoxic T cell degranulation(GO:0043316) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.0 | 0.1 | GO:2000424 | T-helper 1 cell lineage commitment(GO:0002296) negative regulation of complement-dependent cytotoxicity(GO:1903660) regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.0 | 0.1 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:1904884 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.1 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.1 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.0 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.1 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.1 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.0 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.1 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.0 | 0.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.1 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.1 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.0 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.0 | 0.1 | GO:0009227 | nucleotide-sugar catabolic process(GO:0009227) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:1990379 | lipid transport across blood brain barrier(GO:1990379) |
| 0.0 | 0.1 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.0 | 0.0 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.0 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.1 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.1 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:1904640 | response to methionine(GO:1904640) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.0 | GO:0035700 | astrocyte chemotaxis(GO:0035700) regulation of astrocyte chemotaxis(GO:2000458) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.1 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.1 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) |
| 0.0 | 0.4 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.2 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.1 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) |
| 0.0 | 0.0 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.1 | GO:2000535 | exocyst assembly(GO:0001927) entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.0 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.0 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.0 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.1 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.0 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.0 | GO:0033986 | response to methanol(GO:0033986) |
| 0.0 | 0.0 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.0 | 0.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.0 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.1 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 0.0 | 0.0 | GO:1902963 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.1 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.0 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.0 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.1 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031230 | intrinsic component of cell outer membrane(GO:0031230) integral component of cell outer membrane(GO:0045203) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.1 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.0 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.0 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.0 | GO:0005713 | chiasma(GO:0005712) recombination nodule(GO:0005713) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.2 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 0.2 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.1 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) glycoprotein transporter activity(GO:0034437) |
| 0.0 | 0.2 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.2 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.0 | 0.3 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.1 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 0.0 | 0.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.1 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.0 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.0 | GO:0001565 | phorbol ester receptor activity(GO:0001565) |
| 0.0 | 0.1 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.0 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.0 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.0 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.0 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.0 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.0 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |