Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
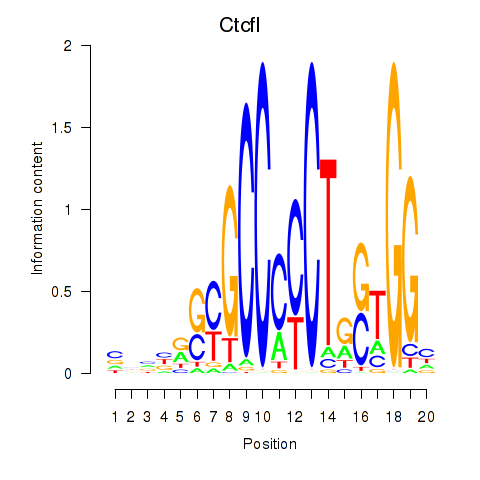
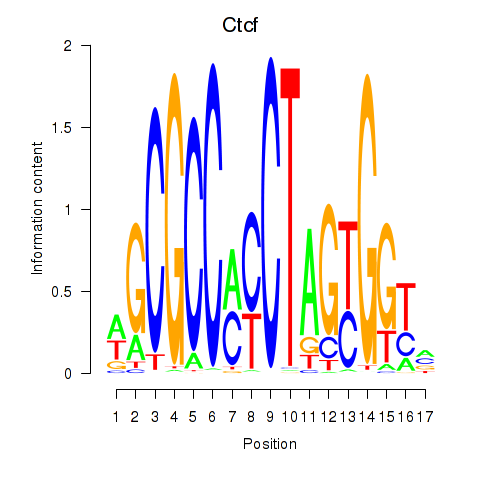
Results for Ctcfl_Ctcf
Z-value: 1.09
Transcription factors associated with Ctcfl_Ctcf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ctcfl
|
ENSRNOG00000028661 | CCCTC-binding factor like |
|
Ctcf
|
ENSRNOG00000017674 | CCCTC-binding factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ctcfl | rn6_v1_chr3_-_171166454_171166454 | 0.29 | 6.3e-01 | Click! |
| Ctcf | rn6_v1_chr19_+_37600148_37600148 | 0.12 | 8.5e-01 | Click! |
Activity profile of Ctcfl_Ctcf motif
Sorted Z-values of Ctcfl_Ctcf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_9695292 | 1.62 |
ENSRNOT00000036162
|
Prr7
|
proline rich 7 (synaptic) |
| chr10_-_15600858 | 0.95 |
ENSRNOT00000035459
|
Hbq1b
|
hemoglobin, theta 1B |
| chr19_+_41482728 | 0.77 |
ENSRNOT00000022943
|
Calb2
|
calbindin 2 |
| chr3_+_164424515 | 0.60 |
ENSRNOT00000083876
|
Cebpb
|
CCAAT/enhancer binding protein beta |
| chr12_+_47551935 | 0.59 |
ENSRNOT00000056932
|
RGD1560398
|
RGD1560398 |
| chr12_+_24651314 | 0.58 |
ENSRNOT00000077016
ENSRNOT00000071569 |
Vps37d
|
VPS37D, ESCRT-I subunit |
| chr16_+_21282467 | 0.47 |
ENSRNOT00000065345
|
Yjefn3
|
YjeF N-terminal domain containing 3 |
| chr6_+_137997335 | 0.47 |
ENSRNOT00000006872
|
Tmem121
|
transmembrane protein 121 |
| chr10_-_90393317 | 0.46 |
ENSRNOT00000028563
|
Fam171a2
|
family with sequence similarity 171, member A2 |
| chr19_-_26094756 | 0.42 |
ENSRNOT00000067780
|
Junb
|
JunB proto-oncogene, AP-1 transcription factor subunit |
| chr17_-_13393243 | 0.40 |
ENSRNOT00000018252
|
Gadd45g
|
growth arrest and DNA-damage-inducible, gamma |
| chr20_-_5618254 | 0.40 |
ENSRNOT00000092326
ENSRNOT00000000576 |
Bak1
|
BCL2-antagonist/killer 1 |
| chr8_-_117237229 | 0.37 |
ENSRNOT00000071381
|
Klhdc8b
|
kelch domain containing 8B |
| chr10_-_85049331 | 0.36 |
ENSRNOT00000012538
|
Tbx21
|
T-box 21 |
| chrX_+_72684329 | 0.35 |
ENSRNOT00000057644
|
Dmrtc1c1
|
DMRT-like family C1c1 |
| chr8_+_71822129 | 0.35 |
ENSRNOT00000089147
|
Dapk2
|
death-associated protein kinase 2 |
| chr1_+_221792221 | 0.34 |
ENSRNOT00000054828
|
Nrxn2
|
neurexin 2 |
| chr10_+_13723405 | 0.32 |
ENSRNOT00000072789
|
Abca3
|
ATP binding cassette subfamily A member 3 |
| chr12_+_48257609 | 0.29 |
ENSRNOT00000031658
ENSRNOT00000090277 |
Alkbh2
|
alkB homolog 2, alpha-ketoglutarate-dependent dioxygenase |
| chr18_-_37776453 | 0.29 |
ENSRNOT00000087876
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr5_-_151397603 | 0.29 |
ENSRNOT00000076866
|
Gpr3
|
G protein-coupled receptor 3 |
| chr16_+_20672374 | 0.28 |
ENSRNOT00000063780
|
RGD1566239
|
similar to RIKEN cDNA 2810428I15 |
| chr15_-_38276159 | 0.27 |
ENSRNOT00000084698
ENSRNOT00000015233 |
Micu2
|
mitochondrial calcium uptake 2 |
| chr3_+_148150698 | 0.27 |
ENSRNOT00000087075
ENSRNOT00000010251 |
Hm13
|
histocompatibility minor 13 |
| chr14_+_83752393 | 0.26 |
ENSRNOT00000081123
|
Selenom
|
selenoprotein M |
| chr3_+_154395187 | 0.26 |
ENSRNOT00000050810
|
Vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr7_+_140781799 | 0.25 |
ENSRNOT00000087932
|
Dnajc22
|
DnaJ heat shock protein family (Hsp40) member C22 |
| chr1_-_142724511 | 0.25 |
ENSRNOT00000014639
|
Nmb
|
neuromedin B |
| chr3_-_154627257 | 0.25 |
ENSRNOT00000018328
|
Tgm2
|
transglutaminase 2 |
| chr6_-_11494459 | 0.25 |
ENSRNOT00000021570
|
Kcnk12
|
potassium two pore domain channel subfamily K member 12 |
| chr4_+_25435873 | 0.25 |
ENSRNOT00000000018
|
Steap1
|
STEAP family member 1 |
| chr10_-_46332172 | 0.24 |
ENSRNOT00000004475
|
Rasd1
|
ras related dexamethasone induced 1 |
| chr11_+_77815181 | 0.24 |
ENSRNOT00000002640
|
Cldn1
|
claudin 1 |
| chr1_-_101131012 | 0.24 |
ENSRNOT00000082283
ENSRNOT00000093498 ENSRNOT00000093559 |
Flt3lg
Rpl13a
|
fms-related tyrosine kinase 3 ligand ribosomal protein L13A |
| chr1_-_52962388 | 0.23 |
ENSRNOT00000033685
|
T2
|
brachyury 2 |
| chr7_-_70452675 | 0.23 |
ENSRNOT00000090498
|
AC114111.1
|
|
| chr11_+_88122271 | 0.21 |
ENSRNOT00000002540
|
Sdf2l1
|
stromal cell-derived factor 2-like 1 |
| chr6_-_128989812 | 0.20 |
ENSRNOT00000085943
|
LOC100909439
|
ankyrin repeat and SOCS box protein 2-like |
| chr16_+_19767264 | 0.20 |
ENSRNOT00000051802
ENSRNOT00000092073 |
Ocel1
|
occludin/ELL domain containing 1 |
| chr2_-_199771896 | 0.20 |
ENSRNOT00000043937
|
Chd1l
|
chromodomain helicase DNA binding protein 1-like |
| chr3_+_72191533 | 0.20 |
ENSRNOT00000044319
|
Ube2l6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr15_-_19575914 | 0.19 |
ENSRNOT00000043897
|
AABR07017253.1
|
|
| chr5_-_61077627 | 0.18 |
ENSRNOT00000015338
|
Shb
|
SH2 domain containing adaptor protein B |
| chr5_+_143081326 | 0.18 |
ENSRNOT00000067204
|
Meaf6
|
MYST/Esa1-associated factor 6 |
| chr10_-_29450644 | 0.18 |
ENSRNOT00000087937
|
Adra1b
|
adrenoceptor alpha 1B |
| chr20_-_10680283 | 0.18 |
ENSRNOT00000001579
|
Sik1
|
salt-inducible kinase 1 |
| chr6_+_136720582 | 0.18 |
ENSRNOT00000086868
|
Kif26a
|
kinesin family member 26A |
| chr11_+_33845463 | 0.18 |
ENSRNOT00000041838
|
Cbr1
|
carbonyl reductase 1 |
| chr2_+_225827504 | 0.17 |
ENSRNOT00000018343
|
Gclm
|
glutamate cysteine ligase, modifier subunit |
| chr7_-_77159457 | 0.17 |
ENSRNOT00000085626
|
Klf10
|
Kruppel-like factor 10 |
| chr10_+_55013703 | 0.17 |
ENSRNOT00000032785
|
Pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr1_+_175445088 | 0.17 |
ENSRNOT00000036718
|
Adm
|
adrenomedullin |
| chr2_+_251817694 | 0.17 |
ENSRNOT00000019964
|
RGD1560065
|
similar to RIKEN cDNA 2410004B18 |
| chr13_-_50509916 | 0.16 |
ENSRNOT00000076747
|
Ren
|
renin |
| chr9_-_43127887 | 0.16 |
ENSRNOT00000021685
|
Ankrd39
|
ankyrin repeat domain 39 |
| chr15_-_29369504 | 0.16 |
ENSRNOT00000060297
|
AABR07017624.1
|
|
| chrX_+_45637415 | 0.16 |
ENSRNOT00000050544
|
RGD1563378
|
similar to ferritin heavy polypeptide-like 17 |
| chr9_+_10941613 | 0.16 |
ENSRNOT00000070794
|
Sema6b
|
semaphorin 6B |
| chr6_+_136720266 | 0.16 |
ENSRNOT00000018278
|
Kif26a
|
kinesin family member 26A |
| chr12_+_23151180 | 0.16 |
ENSRNOT00000059486
|
Cux1
|
cut-like homeobox 1 |
| chr7_+_12022285 | 0.15 |
ENSRNOT00000024080
|
Rexo1
|
RNA exonuclease 1 homolog |
| chr7_-_12424367 | 0.15 |
ENSRNOT00000060698
|
Midn
|
midnolin |
| chr7_+_2875909 | 0.15 |
ENSRNOT00000028244
|
Smarcc2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
| chr20_+_5050327 | 0.15 |
ENSRNOT00000083353
|
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr1_-_254671596 | 0.15 |
ENSRNOT00000025450
|
Htr7
|
5-hydroxytryptamine receptor 7 |
| chr5_-_156141537 | 0.15 |
ENSRNOT00000019004
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr7_-_11513415 | 0.15 |
ENSRNOT00000082486
|
Thop1
|
thimet oligopeptidase 1 |
| chr3_+_160231914 | 0.15 |
ENSRNOT00000014411
|
Kcnk15
|
potassium two pore domain channel subfamily K member 15 |
| chr5_-_157368450 | 0.14 |
ENSRNOT00000023143
ENSRNOT00000079855 |
Otud3
|
OTU deubiquitinase 3 |
| chr16_+_9563218 | 0.14 |
ENSRNOT00000035915
|
Arhgap22
|
Rho GTPase activating protein 22 |
| chr1_+_82480195 | 0.14 |
ENSRNOT00000028051
|
Tgfb1
|
transforming growth factor, beta 1 |
| chrX_-_14890606 | 0.14 |
ENSRNOT00000049864
|
RGD1560784
|
similar to RIKEN cDNA B630019K06 |
| chr7_-_2909144 | 0.14 |
ENSRNOT00000082518
ENSRNOT00000089074 ENSRNOT00000085644 |
Myl6
|
myosin light chain 6 |
| chr8_-_50526843 | 0.14 |
ENSRNOT00000092188
|
AABR07070085.1
|
|
| chr9_+_97355924 | 0.14 |
ENSRNOT00000026558
|
Ackr3
|
atypical chemokine receptor 3 |
| chr3_+_170252901 | 0.14 |
ENSRNOT00000005871
|
Mc3r
|
melanocortin 3 receptor |
| chr9_-_92775816 | 0.14 |
ENSRNOT00000029635
|
RGD1563917
|
similar to Nuclear autoantigen Sp-100 (Speckled 100 kDa) |
| chr7_-_2825498 | 0.14 |
ENSRNOT00000086656
ENSRNOT00000031362 |
Nabp2
|
nucleic acid binding protein 2 |
| chr7_-_142260896 | 0.14 |
ENSRNOT00000073536
|
Smagp
|
small cell adhesion glycoprotein |
| chr5_+_152606847 | 0.14 |
ENSRNOT00000089070
|
Pafah2
|
platelet-activating factor acetylhydrolase 2 |
| chr19_+_10519493 | 0.13 |
ENSRNOT00000030229
|
Ccdc102a
|
coiled-coil domain containing 102A |
| chr14_-_84170301 | 0.13 |
ENSRNOT00000080413
|
Slc35e4
|
solute carrier family 35, member E4 |
| chr16_-_20890949 | 0.13 |
ENSRNOT00000081977
|
Homer3
|
homer scaffolding protein 3 |
| chr8_-_62424303 | 0.13 |
ENSRNOT00000091223
|
Csk
|
c-src tyrosine kinase |
| chr19_+_26106838 | 0.13 |
ENSRNOT00000035987
|
Hook2
|
hook microtubule-tethering protein 2 |
| chr1_-_80666566 | 0.13 |
ENSRNOT00000082125
ENSRNOT00000025388 |
Nectin2
|
nectin cell adhesion molecule 2 |
| chr15_+_31948035 | 0.13 |
ENSRNOT00000071627
|
AABR07017868.1
|
|
| chr3_+_160852164 | 0.13 |
ENSRNOT00000019127
|
Rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr9_-_15306465 | 0.13 |
ENSRNOT00000019404
|
Frs3
|
fibroblast growth factor receptor substrate 3 |
| chr20_+_3995544 | 0.12 |
ENSRNOT00000000527
|
Tap2
|
transporter 2, ATP binding cassette subfamily B member |
| chr7_-_58587787 | 0.12 |
ENSRNOT00000005814
|
Lgr5
|
leucine rich repeat containing G protein coupled receptor 5 |
| chr1_-_254671778 | 0.12 |
ENSRNOT00000025493
|
Htr7
|
5-hydroxytryptamine receptor 7 |
| chr10_-_68517564 | 0.12 |
ENSRNOT00000086961
|
Asic2
|
acid sensing ion channel subunit 2 |
| chr10_-_14092289 | 0.12 |
ENSRNOT00000019624
|
Ndufb10
|
NADH:ubiquinone oxidoreductase subunit B10 |
| chr7_-_138707221 | 0.12 |
ENSRNOT00000009199
|
Amigo2
|
adhesion molecule with Ig like domain 2 |
| chr18_+_30527705 | 0.12 |
ENSRNOT00000027168
|
Pcdhb14
|
protocadherin beta 14 |
| chr10_+_86950557 | 0.12 |
ENSRNOT00000014153
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr14_+_82769642 | 0.12 |
ENSRNOT00000065393
|
Ctbp1
|
C-terminal binding protein 1 |
| chr2_+_198772937 | 0.11 |
ENSRNOT00000028812
|
Itga10
|
integrin subunit alpha 10 |
| chr2_-_188471988 | 0.11 |
ENSRNOT00000027785
|
Hcn3
|
hyperpolarization-activated cyclic nucleotide-gated potassium channel 3 |
| chr1_+_173532803 | 0.11 |
ENSRNOT00000021017
|
Eif3f
|
eukaryotic translation initiation factor 3, subunit F |
| chr7_-_11513581 | 0.11 |
ENSRNOT00000027045
|
Thop1
|
thimet oligopeptidase 1 |
| chr17_+_70684340 | 0.11 |
ENSRNOT00000051067
|
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr5_-_159602251 | 0.11 |
ENSRNOT00000011394
|
Necap2
|
NECAP endocytosis associated 2 |
| chr10_+_105861743 | 0.10 |
ENSRNOT00000064410
|
Mgat5b
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B |
| chr10_+_40543288 | 0.10 |
ENSRNOT00000016755
|
Slc36a1
|
solute carrier family 36 member 1 |
| chr2_-_187133993 | 0.10 |
ENSRNOT00000019502
|
Pear1
|
platelet endothelial aggregation receptor 1 |
| chr12_+_13299859 | 0.10 |
ENSRNOT00000039191
|
LOC683674
|
similar to Protein C7orf26 homolog |
| chr10_-_109757550 | 0.10 |
ENSRNOT00000054957
|
Arhgdia
|
Rho GDP dissociation inhibitor alpha |
| chrX_-_45522665 | 0.10 |
ENSRNOT00000030771
|
RGD1562200
|
similar to GS2 gene |
| chr15_-_23580342 | 0.10 |
ENSRNOT00000013331
ENSRNOT00000085767 |
Cnih1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr15_-_33193537 | 0.10 |
ENSRNOT00000016401
|
Haus4
|
HAUS augmin-like complex, subunit 4 |
| chr17_+_26785029 | 0.10 |
ENSRNOT00000022065
|
Eef1e1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr9_-_10441834 | 0.10 |
ENSRNOT00000043704
|
Rpl36
|
ribosomal protein L36 |
| chr11_+_73738433 | 0.10 |
ENSRNOT00000002353
|
Tmem44
|
transmembrane protein 44 |
| chr6_-_46631983 | 0.10 |
ENSRNOT00000045963
|
Sox11
|
SRY box 11 |
| chr19_+_37652969 | 0.10 |
ENSRNOT00000041970
|
Carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr20_-_5212624 | 0.10 |
ENSRNOT00000074261
|
LOC103689996
|
antigen peptide transporter 2 |
| chr15_-_34444244 | 0.09 |
ENSRNOT00000027612
|
Cideb
|
cell death-inducing DFFA-like effector b |
| chr11_+_36851038 | 0.09 |
ENSRNOT00000002221
ENSRNOT00000061047 |
Pcp4
|
Purkinje cell protein 4 |
| chr18_+_30509393 | 0.09 |
ENSRNOT00000043846
|
Pcdhb12
|
protocadherin beta 12 |
| chr18_-_57245666 | 0.09 |
ENSRNOT00000080365
|
Ablim3
|
actin binding LIM protein family, member 3 |
| chr13_-_53108713 | 0.09 |
ENSRNOT00000035404
|
RGD1311892
|
similar to hypothetical protein FLJ10901 |
| chr3_-_11382004 | 0.09 |
ENSRNOT00000047921
ENSRNOT00000064039 |
Dnm1
|
dynamin 1 |
| chr10_-_86393141 | 0.09 |
ENSRNOT00000009485
|
Mien1
|
migration and invasion enhancer 1 |
| chr5_-_164927869 | 0.09 |
ENSRNOT00000012080
|
Draxin
|
dorsal inhibitory axon guidance protein |
| chr3_-_36660758 | 0.09 |
ENSRNOT00000006111
|
Rnd3
|
Rho family GTPase 3 |
| chr1_+_47605262 | 0.09 |
ENSRNOT00000089458
|
Fndc1
|
fibronectin type III domain containing 1 |
| chr10_+_66099531 | 0.09 |
ENSRNOT00000056192
|
Lyrm9
|
LYR motif containing 9 |
| chr4_+_121612332 | 0.09 |
ENSRNOT00000077374
ENSRNOT00000084494 |
Txnrd3
|
thioredoxin reductase 3 |
| chr2_+_157854667 | 0.09 |
ENSRNOT00000039294
|
AC119603.1
|
|
| chr6_-_126582034 | 0.09 |
ENSRNOT00000010656
ENSRNOT00000080829 |
Itpk1
|
inositol-tetrakisphosphate 1-kinase |
| chr7_-_144272578 | 0.09 |
ENSRNOT00000020676
|
Atp5g2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr8_-_94120433 | 0.09 |
ENSRNOT00000080195
ENSRNOT00000014600 |
Ube3d
|
ubiquitin protein ligase E3D |
| chr3_+_168124673 | 0.09 |
ENSRNOT00000070809
|
Pfdn4
|
prefoldin subunit 4 |
| chr2_+_195719543 | 0.09 |
ENSRNOT00000028324
|
Celf3
|
CUGBP, Elav-like family member 3 |
| chr1_-_15374850 | 0.09 |
ENSRNOT00000016728
|
Pex7
|
peroxisomal biogenesis factor 7 |
| chr1_-_85517360 | 0.08 |
ENSRNOT00000026114
|
Eid2
|
EP300 interacting inhibitor of differentiation 2 |
| chr11_-_67756799 | 0.08 |
ENSRNOT00000030975
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr20_-_5244386 | 0.08 |
ENSRNOT00000070886
|
RT1-DMa
|
RT1 class II, locus DMa |
| chr3_+_175548174 | 0.08 |
ENSRNOT00000091941
|
Adrm1
|
adhesion regulating molecule 1 |
| chr3_-_148313810 | 0.08 |
ENSRNOT00000010762
|
Bcl2l1
|
Bcl2-like 1 |
| chr7_-_12793711 | 0.08 |
ENSRNOT00000013762
|
Palm
|
paralemmin |
| chr17_+_31493107 | 0.08 |
ENSRNOT00000023611
ENSRNOT00000086264 |
Tubb2a
|
tubulin, beta 2A class IIa |
| chr12_+_13716596 | 0.08 |
ENSRNOT00000080216
|
Actb
|
actin, beta |
| chr20_+_7818289 | 0.08 |
ENSRNOT00000042539
|
Ppard
|
peroxisome proliferator-activated receptor delta |
| chr1_-_89483988 | 0.08 |
ENSRNOT00000028603
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7 |
| chr9_-_121972055 | 0.08 |
ENSRNOT00000089735
|
AABR07068851.1
|
clusterin-like protein 1 |
| chr7_-_11963268 | 0.08 |
ENSRNOT00000080342
ENSRNOT00000025266 |
Csnk1g2
|
casein kinase 1, gamma 2 |
| chr3_+_164274710 | 0.08 |
ENSRNOT00000012939
|
Snai1
|
snail family transcriptional repressor 1 |
| chr4_-_56453563 | 0.08 |
ENSRNOT00000060362
|
Prrt4
|
proline-rich transmembrane protein 4 |
| chr10_-_88266210 | 0.08 |
ENSRNOT00000090702
ENSRNOT00000020603 |
Hap1
|
huntingtin-associated protein 1 |
| chr2_-_210738378 | 0.08 |
ENSRNOT00000025746
|
Gstm6
|
glutathione S-transferase, mu 6 |
| chr13_+_79886832 | 0.08 |
ENSRNOT00000077069
ENSRNOT00000035558 ENSRNOT00000076417 |
Pigc
|
phosphatidylinositol glycan anchor biosynthesis, class C |
| chr8_+_117920944 | 0.08 |
ENSRNOT00000028125
|
Nme6
|
NME/NM23 nucleoside diphosphate kinase 6 |
| chr4_+_155313671 | 0.08 |
ENSRNOT00000020812
|
Mfap5
|
microfibrillar associated protein 5 |
| chrX_-_104726816 | 0.07 |
ENSRNOT00000005165
|
Tspan6
|
tetraspanin 6 |
| chr7_-_125407806 | 0.07 |
ENSRNOT00000084666
|
RGD1566029
|
similar to mKIAA1644 protein |
| chr2_+_189581644 | 0.07 |
ENSRNOT00000092112
|
Rab13
|
RAB13, member RAS oncogene family |
| chr9_+_40975836 | 0.07 |
ENSRNOT00000084470
|
Ptpn18
|
protein tyrosine phosphatase, non-receptor type 18 |
| chr7_-_11699436 | 0.07 |
ENSRNOT00000049221
ENSRNOT00000066037 |
Tmprss9
|
transmembrane protease, serine 9 |
| chr3_+_2642531 | 0.07 |
ENSRNOT00000081798
|
Fut7
|
fucosyltransferase 7 |
| chr1_-_226924244 | 0.07 |
ENSRNOT00000028971
|
Tmem132a
|
transmembrane protein 132A |
| chrX_+_63343546 | 0.07 |
ENSRNOT00000076315
|
Klhl15
|
kelch-like family member 15 |
| chr13_-_110257367 | 0.07 |
ENSRNOT00000005576
|
Dtl
|
denticleless E3 ubiquitin protein ligase homolog |
| chr10_+_108395860 | 0.07 |
ENSRNOT00000075796
|
Gaa
|
glucosidase, alpha, acid |
| chrX_-_105568343 | 0.07 |
ENSRNOT00000029807
|
Armcx6
|
armadillo repeat containing, X-linked 6 |
| chr6_-_43448280 | 0.07 |
ENSRNOT00000081110
|
Adam17
|
ADAM metallopeptidase domain 17 |
| chr20_-_3374344 | 0.07 |
ENSRNOT00000082999
|
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr1_-_79930263 | 0.07 |
ENSRNOT00000019219
|
Foxa3
|
forkhead box A3 |
| chr3_+_103747654 | 0.07 |
ENSRNOT00000006887
|
Nop10
|
NOP10 ribonucleoprotein |
| chr1_-_77865870 | 0.07 |
ENSRNOT00000017353
|
Ehd2
|
EH-domain containing 2 |
| chr9_-_71313940 | 0.07 |
ENSRNOT00000019670
|
Mettl21a
|
methyltransferase like 21A |
| chr1_+_222519615 | 0.07 |
ENSRNOT00000083585
|
Rcor2
|
REST corepressor 2 |
| chr20_+_6869767 | 0.07 |
ENSRNOT00000000631
ENSRNOT00000086330 ENSRNOT00000093188 ENSRNOT00000093736 |
RGD735065
|
similar to GI:13385412-like protein splice form I |
| chr10_+_92289107 | 0.06 |
ENSRNOT00000050070
|
Mapt
|
microtubule-associated protein tau |
| chr20_+_3935529 | 0.06 |
ENSRNOT00000072458
|
LOC108348072
|
class II histocompatibility antigen, M alpha chain |
| chr12_+_2228670 | 0.06 |
ENSRNOT00000001327
|
Trappc5
|
trafficking protein particle complex 5 |
| chr19_+_46761570 | 0.06 |
ENSRNOT00000058779
|
Wwox
|
WW domain-containing oxidoreductase |
| chr17_+_9631925 | 0.06 |
ENSRNOT00000018114
|
Ddx41
|
DEAD-box helicase 41 |
| chr19_+_38397466 | 0.06 |
ENSRNOT00000034386
|
Nob1
|
NIN1/PSMD8 binding protein 1 homolog |
| chr1_+_101213788 | 0.06 |
ENSRNOT00000090838
|
Tead2
|
TEA domain transcription factor 2 |
| chr15_+_45712821 | 0.06 |
ENSRNOT00000083381
ENSRNOT00000045833 |
Fam124a
|
family with sequence similarity 124 member A |
| chr10_-_104358253 | 0.06 |
ENSRNOT00000005942
|
Caskin2
|
cask-interacting protein 2 |
| chr1_+_40389638 | 0.06 |
ENSRNOT00000021471
|
Plekhg1
|
pleckstrin homology and RhoGEF domain containing G1 |
| chr6_+_51662224 | 0.06 |
ENSRNOT00000060006
|
Ccdc71l
|
coiled-coil domain containing 71-like |
| chr7_+_94777702 | 0.06 |
ENSRNOT00000049001
|
H3f3c
|
H3 histone, family 3C |
| chr10_+_11877903 | 0.06 |
ENSRNOT00000076532
ENSRNOT00000075890 |
RGD1561796
|
RGD1561796 |
| chr20_+_28920616 | 0.06 |
ENSRNOT00000070897
|
P4ha1
|
prolyl 4-hydroxylase subunit alpha 1 |
| chr13_-_99101208 | 0.06 |
ENSRNOT00000004329
|
H3f3a
|
H3 histone family member 3A |
| chr6_-_29483460 | 0.06 |
ENSRNOT00000007160
|
Klhl29
|
kelch-like family member 29 |
| chr7_-_23843505 | 0.06 |
ENSRNOT00000006366
ENSRNOT00000077577 |
Fbxo7
|
F-box protein 7 |
| chr1_-_198298076 | 0.06 |
ENSRNOT00000027033
|
Ino80e
|
INO80 complex subunit E |
| chr19_+_10119253 | 0.06 |
ENSRNOT00000017971
|
Zfp319
|
zinc finger protein 319 |
| chr18_+_29993361 | 0.06 |
ENSRNOT00000075810
|
Pcdha4
|
protocadherin alpha 4 |
| chr10_+_35392762 | 0.06 |
ENSRNOT00000059277
|
Rasgef1c
|
RasGEF domain family, member 1C |
| chr4_-_171176581 | 0.06 |
ENSRNOT00000030850
|
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor |
| chr1_-_101131413 | 0.06 |
ENSRNOT00000093729
|
Flt3lg
|
fms-related tyrosine kinase 3 ligand |
| chr9_+_11031841 | 0.06 |
ENSRNOT00000074695
|
Sh3gl1
|
SH3 domain-containing GRB2-like 1 |
| chr15_+_19429508 | 0.06 |
ENSRNOT00000047113
|
LOC100361240
|
ribosomal protein S10-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ctcfl_Ctcf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0035711 | granuloma formation(GO:0002432) T-helper 1 cell activation(GO:0035711) |
| 0.1 | 0.3 | GO:2000424 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.1 | 0.3 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.3 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.2 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.1 | 0.3 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 0.2 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 0.2 | GO:1903544 | positive regulation of bicellular tight junction assembly(GO:1903348) response to butyrate(GO:1903544) |
| 0.1 | 0.3 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) |
| 0.1 | 0.3 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 0.4 | GO:0002339 | B cell selection(GO:0002339) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 0.2 | GO:1901662 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 | 0.3 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.1 | 0.3 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 | 0.4 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.1 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.2 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) |
| 0.0 | 0.1 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 0.1 | GO:0002489 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) cytosol to ER transport(GO:0046967) peptide antigen transport(GO:0046968) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.2 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.2 | GO:0002003 | angiotensin maturation(GO:0002003) renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.3 | GO:0035934 | corticosterone secretion(GO:0035934) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.2 | GO:0097461 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.1 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.0 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.1 | GO:1905218 | cellular response to astaxanthin(GO:1905218) |
| 0.0 | 0.1 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.0 | 0.2 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.1 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.1 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.0 | 0.1 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.0 | 0.1 | GO:1904009 | cellular response to monosodium glutamate(GO:1904009) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:1902164 | positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.3 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.1 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.0 | 0.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) |
| 0.0 | 0.1 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.0 | 0.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.0 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.0 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 | 0.2 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.1 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.0 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.0 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.1 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.2 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:0070778 | L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 0.0 | 0.0 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.0 | 0.1 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.0 | 0.1 | GO:0006527 | citrulline metabolic process(GO:0000052) arginine catabolic process(GO:0006527) |
| 0.0 | 0.0 | GO:0015942 | formate metabolic process(GO:0015942) |
| 0.0 | 0.1 | GO:1903350 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.1 | 0.3 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.1 | 0.4 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.2 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.3 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.8 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.2 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0033290 | eukaryotic 43S preinitiation complex(GO:0016282) eukaryotic 48S preinitiation complex(GO:0033290) eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.0 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.0 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.0 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.1 | 0.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.3 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.2 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.4 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.3 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.0 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.0 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.6 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |