Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Crx_Gsc
Z-value: 0.52
Transcription factors associated with Crx_Gsc
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
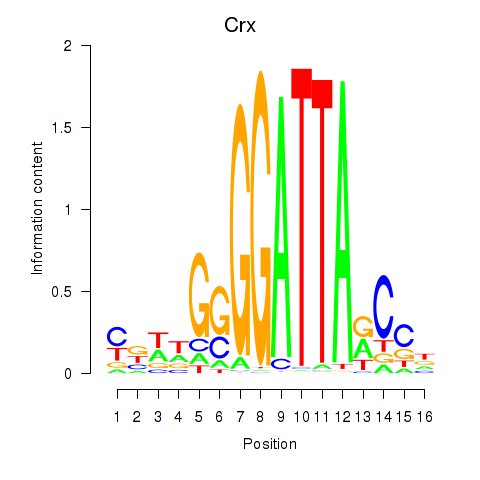
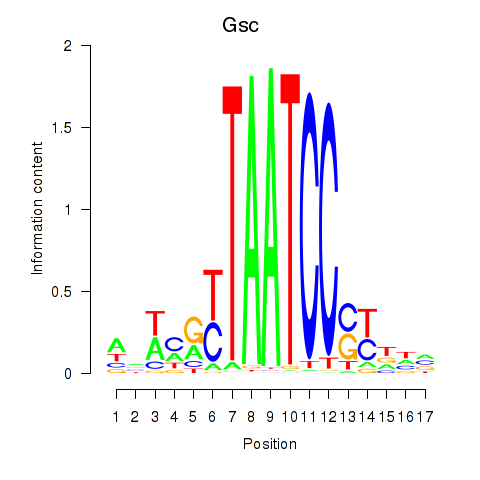
Crx
|
ENSRNOG00000013890 | cone-rod homeobox |
|
Gsc
|
ENSRNOG00000010605 | goosecoid homeobox |
Activity profile of Crx_Gsc motif
Sorted Z-values of Crx_Gsc motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_55050284 | 0.32 |
ENSRNOT00000013242
|
Pih1d2
|
PIH1 domain containing 2 |
| chr3_+_138174054 | 0.25 |
ENSRNOT00000007946
|
Banf2
|
barrier to autointegration factor 2 |
| chr5_-_135025084 | 0.17 |
ENSRNOT00000018766
|
Tspan1
|
tetraspanin 1 |
| chr3_-_40477754 | 0.15 |
ENSRNOT00000091168
|
AABR07052166.1
|
|
| chr2_+_206342066 | 0.15 |
ENSRNOT00000026556
|
Ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 |
| chr20_+_4967194 | 0.15 |
ENSRNOT00000070846
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr10_+_70884531 | 0.14 |
ENSRNOT00000015199
|
Ccl4
|
C-C motif chemokine ligand 4 |
| chr10_+_15156207 | 0.13 |
ENSRNOT00000020363
|
Ccdc78
|
coiled-coil domain containing 78 |
| chr5_+_138300107 | 0.12 |
ENSRNOT00000047151
|
Cldn19
|
claudin 19 |
| chr10_+_109774878 | 0.11 |
ENSRNOT00000054954
|
Anapc11
|
anaphase promoting complex subunit 11 |
| chr4_+_94696965 | 0.11 |
ENSRNOT00000064696
|
Grid2
|
glutamate ionotropic receptor delta type subunit 2 |
| chr4_+_81244469 | 0.11 |
ENSRNOT00000044407
ENSRNOT00000015828 |
Cbx3
|
chromobox 3 |
| chr7_-_13751271 | 0.11 |
ENSRNOT00000009931
|
Slc1a6
|
solute carrier family 1 member 6 |
| chrX_-_37003642 | 0.10 |
ENSRNOT00000040770
ENSRNOT00000058834 ENSRNOT00000058833 |
Adgrg2
|
adhesion G protein-coupled receptor G2 |
| chr4_-_170912629 | 0.10 |
ENSRNOT00000055691
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr10_+_39066716 | 0.10 |
ENSRNOT00000010729
|
Il5
|
interleukin 5 |
| chr13_-_111474411 | 0.10 |
ENSRNOT00000072729
|
Hhat
|
hedgehog acyltransferase |
| chr6_-_91581262 | 0.10 |
ENSRNOT00000034507
|
AABR07064716.1
|
|
| chr17_+_72429618 | 0.10 |
ENSRNOT00000026187
|
Gata3
|
GATA binding protein 3 |
| chr17_+_18031228 | 0.09 |
ENSRNOT00000081708
|
Tpmt
|
thiopurine S-methyltransferase |
| chrM_+_7919 | 0.09 |
ENSRNOT00000046108
|
Mt-atp6
|
mitochondrially encoded ATP synthase 6 |
| chr10_+_54512983 | 0.09 |
ENSRNOT00000005204
|
Stx8
|
syntaxin 8 |
| chr14_-_100184192 | 0.09 |
ENSRNOT00000007044
|
Plek
|
pleckstrin |
| chr17_-_2705123 | 0.09 |
ENSRNOT00000024940
|
Olr1652
|
olfactory receptor 1652 |
| chr5_+_103024376 | 0.09 |
ENSRNOT00000058536
|
AABR07049064.1
|
|
| chr10_+_14105750 | 0.09 |
ENSRNOT00000090552
|
Msrb1
|
methionine sulfoxide reductase B1 |
| chrX_-_123731294 | 0.09 |
ENSRNOT00000092574
ENSRNOT00000032618 |
Upf3b
|
UPF3 regulator of nonsense transcripts homolog B (yeast) |
| chr1_-_206394346 | 0.08 |
ENSRNOT00000038122
|
LOC100302465
|
hypothetical LOC100302465 |
| chr17_-_43776460 | 0.08 |
ENSRNOT00000089055
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chr10_-_85725429 | 0.08 |
ENSRNOT00000005471
|
Rpl23
|
ribosomal protein L23 |
| chr15_+_34452116 | 0.08 |
ENSRNOT00000027647
|
Ltb4r
|
leukotriene B4 receptor |
| chrM_+_7758 | 0.08 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr5_+_135997052 | 0.08 |
ENSRNOT00000024921
|
Tctex1d4
|
Tctex1 domain containing 4 |
| chr1_+_100393303 | 0.08 |
ENSRNOT00000026251
|
Syt3
|
synaptotagmin 3 |
| chr9_+_95256627 | 0.08 |
ENSRNOT00000025291
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr2_+_240461505 | 0.07 |
ENSRNOT00000019507
|
Bdh2
|
3-hydroxybutyrate dehydrogenase 2 |
| chr7_+_114323542 | 0.07 |
ENSRNOT00000012121
|
Chrac1
|
chromatin accessibility complex 1 |
| chr10_+_104368247 | 0.07 |
ENSRNOT00000006519
|
Llgl2
|
LLGL2, scribble cell polarity complex component |
| chr2_+_58724855 | 0.07 |
ENSRNOT00000089609
|
Capsl
|
calcyphosine-like |
| chr5_-_146795866 | 0.07 |
ENSRNOT00000065640
|
Tlr12
|
toll-like receptor 12 |
| chr7_+_11677340 | 0.07 |
ENSRNOT00000026648
|
Timm13
|
translocase of inner mitochondrial membrane 13 |
| chr15_-_44442875 | 0.07 |
ENSRNOT00000017939
|
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr10_+_72272248 | 0.07 |
ENSRNOT00000003908
|
Car4
|
carbonic anhydrase 4 |
| chr5_-_79008363 | 0.07 |
ENSRNOT00000010040
|
Kif12
|
kinesin family member 12 |
| chr3_-_110964452 | 0.07 |
ENSRNOT00000016356
|
Rmdn3
|
regulator of microtubule dynamics 3 |
| chr10_-_87195075 | 0.06 |
ENSRNOT00000014851
|
Krt24
|
keratin 24 |
| chr4_-_30276372 | 0.06 |
ENSRNOT00000011823
|
Pon1
|
paraoxonase 1 |
| chr14_-_114047527 | 0.06 |
ENSRNOT00000083199
|
AABR07016779.1
|
|
| chr6_-_127534247 | 0.06 |
ENSRNOT00000012500
|
Serpina6
|
serpin family A member 6 |
| chr4_-_51199570 | 0.06 |
ENSRNOT00000010788
|
Slc13a1
|
solute carrier family 13 member 1 |
| chr18_+_36829062 | 0.06 |
ENSRNOT00000025467
ENSRNOT00000086979 |
Tcerg1
|
transcription elongation regulator 1 |
| chr1_+_141447022 | 0.06 |
ENSRNOT00000020690
|
AC096024.1
|
|
| chr17_-_32558180 | 0.06 |
ENSRNOT00000022681
|
Serpinb1b
|
serine (or cysteine) peptidase inhibitor, clade B, member 1b |
| chr3_+_159936856 | 0.06 |
ENSRNOT00000078703
|
Hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr17_-_44744902 | 0.06 |
ENSRNOT00000085381
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr20_+_14577166 | 0.06 |
ENSRNOT00000085583
|
Rtdr1
|
rhabdoid tumor deletion region gene 1 |
| chr14_-_15258207 | 0.06 |
ENSRNOT00000039383
|
Cxcl13
|
C-X-C motif chemokine ligand 13 |
| chrX_+_13441558 | 0.06 |
ENSRNOT00000030170
|
RGD1561661
|
similar to Ferritin light chain (Ferritin L subunit) |
| chr1_+_78659435 | 0.06 |
ENSRNOT00000071271
|
Ceacam9
|
carcinoembryonic antigen-related cell adhesion molecule 9 |
| chr14_+_91557601 | 0.06 |
ENSRNOT00000038733
|
RGD1309870
|
hypothetical LOC289778 |
| chr18_+_40962146 | 0.06 |
ENSRNOT00000035656
|
Arl14epl
|
ADP ribosylation factor like GTPase 14 effector protein like |
| chr7_-_28932641 | 0.06 |
ENSRNOT00000059487
|
Dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr10_+_86303727 | 0.06 |
ENSRNOT00000037752
|
Ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr1_-_38538987 | 0.06 |
ENSRNOT00000090406
ENSRNOT00000071275 |
LOC102551340
|
zinc finger protein 728-like |
| chr19_-_56311991 | 0.06 |
ENSRNOT00000014090
|
Dbndd1
|
dysbindin domain containing 1 |
| chr2_-_115836846 | 0.06 |
ENSRNOT00000014359
|
Cldn11
|
claudin 11 |
| chr19_+_10731855 | 0.06 |
ENSRNOT00000022277
|
Pllp
|
plasmolipin |
| chr2_-_235852708 | 0.06 |
ENSRNOT00000009046
ENSRNOT00000005772 |
Rpl34
|
ribosomal protein L34 |
| chr19_+_51985170 | 0.06 |
ENSRNOT00000019443
|
Hsbp1
|
heat shock factor binding protein 1 |
| chr1_+_216191886 | 0.05 |
ENSRNOT00000054863
|
Tspan32
|
tetraspanin 32 |
| chr1_+_266781617 | 0.05 |
ENSRNOT00000027417
|
Ina
|
internexin neuronal intermediate filament protein, alpha |
| chr3_+_35271786 | 0.05 |
ENSRNOT00000065989
|
Lypd6b
|
LY6/PLAUR domain containing 6B |
| chr14_-_72025137 | 0.05 |
ENSRNOT00000080604
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr14_+_110676090 | 0.05 |
ENSRNOT00000029513
|
Fancl
|
Fanconi anemia, complementation group L |
| chr16_+_9074033 | 0.05 |
ENSRNOT00000027193
|
Fam170b
|
family with sequence similarity 170, member B |
| chr7_-_116955148 | 0.05 |
ENSRNOT00000087328
|
Pycrl
|
pyrroline-5-carboxylate reductase-like |
| chr1_-_253186695 | 0.05 |
ENSRNOT00000080928
|
AC096809.1
|
|
| chr17_-_8980571 | 0.05 |
ENSRNOT00000071426
|
Pcbd2
|
pterin-4 alpha-carbinolamine dehydratase 2 |
| chr5_-_172307431 | 0.05 |
ENSRNOT00000018453
|
Fam213b
|
family with sequence similarity 213, member B |
| chrX_-_14910727 | 0.05 |
ENSRNOT00000085079
ENSRNOT00000030146 |
Ssx1
|
SSX family member 1 |
| chr11_+_87220618 | 0.05 |
ENSRNOT00000041975
|
Gsc2
|
goosecoid homeobox 2 |
| chr3_-_105470475 | 0.05 |
ENSRNOT00000011078
|
Gjd2
|
gap junction protein, delta 2 |
| chr10_+_65586504 | 0.05 |
ENSRNOT00000015535
ENSRNOT00000046388 |
Aldoc
|
aldolase, fructose-bisphosphate C |
| chr9_-_94192813 | 0.05 |
ENSRNOT00000025978
|
Alpp
|
alkaline phosphatase, placental |
| chr19_+_9587653 | 0.05 |
ENSRNOT00000015956
|
Got2
|
glutamic-oxaloacetic transaminase 2 |
| chr2_-_198458041 | 0.05 |
ENSRNOT00000082450
|
Fcgr1a
|
Fc fragment of IgG receptor Ia |
| chr19_+_10596960 | 0.05 |
ENSRNOT00000021769
|
Ciapin1
|
cytokine induced apoptosis inhibitor 1 |
| chr6_-_10899200 | 0.05 |
ENSRNOT00000089104
|
Mcfd2
|
multiple coagulation factor deficiency 2 |
| chr11_-_70211701 | 0.05 |
ENSRNOT00000076114
|
Muc13
|
mucin 13, cell surface associated |
| chr8_+_33514042 | 0.05 |
ENSRNOT00000081614
ENSRNOT00000081525 |
Kcnj1
|
potassium voltage-gated channel subfamily J member 1 |
| chr5_+_98387291 | 0.05 |
ENSRNOT00000046503
|
Tyrp1
|
tyrosinase-related protein 1 |
| chr1_-_1889236 | 0.05 |
ENSRNOT00000087944
|
AABR07000159.2
|
|
| chr6_+_76041669 | 0.05 |
ENSRNOT00000039829
|
Fam177a1
|
family with sequence similarity 177, member A1 |
| chr10_-_109774639 | 0.05 |
ENSRNOT00000054955
|
Alyref
|
Aly/REF export factor |
| chr2_+_187415262 | 0.05 |
ENSRNOT00000056923
|
Gpatch4
|
G patch domain containing 4 |
| chr19_+_10827371 | 0.05 |
ENSRNOT00000024120
|
Fam192a
|
family with sequence similarity 192, member A |
| chr1_+_141832774 | 0.05 |
ENSRNOT00000073302
|
Fuom
|
fucose mutarotase |
| chr4_-_147163467 | 0.05 |
ENSRNOT00000010748
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr3_-_148104483 | 0.05 |
ENSRNOT00000055406
|
Defb25
|
defensin beta 25 |
| chrX_+_70461718 | 0.05 |
ENSRNOT00000078233
ENSRNOT00000003789 |
Kif4a
|
kinesin family member 4A |
| chr15_+_86153628 | 0.04 |
ENSRNOT00000012842
|
Uchl3
|
ubiquitin C-terminal hydrolase L3 |
| chr5_-_57267002 | 0.04 |
ENSRNOT00000011455
|
Bag1
|
Bcl2 associated athanogene 1 |
| chr19_+_37282018 | 0.04 |
ENSRNOT00000021723
|
Tmem208
|
transmembrane protein 208 |
| chr1_-_276012351 | 0.04 |
ENSRNOT00000045642
|
AABR07007000.1
|
|
| chr7_-_64251110 | 0.04 |
ENSRNOT00000006180
|
Srgap1
|
SLIT-ROBO Rho GTPase activating protein 1 |
| chr5_-_32956159 | 0.04 |
ENSRNOT00000078264
|
Cnbd1
|
cyclic nucleotide binding domain containing 1 |
| chr2_+_92573004 | 0.04 |
ENSRNOT00000065774
|
tGap1
|
GTPase activating protein testicular GAP1 |
| chrX_+_124894466 | 0.04 |
ENSRNOT00000080894
|
Mcts1
|
MCTS1, re-initiation and release factor |
| chr2_-_9504134 | 0.04 |
ENSRNOT00000076996
|
Adgrv1
|
adhesion G protein-coupled receptor V1 |
| chr4_-_162726628 | 0.04 |
ENSRNOT00000073877
|
LOC100911272
|
killer cell lectin-like receptor 5-like |
| chr3_-_72071895 | 0.04 |
ENSRNOT00000082857
|
Selenoh
|
selenoprotein H |
| chr11_-_43099412 | 0.04 |
ENSRNOT00000002281
|
Gabrr3
|
gamma-aminobutyric acid type A receptor rho3 subunit |
| chrX_+_105911925 | 0.04 |
ENSRNOT00000052422
|
LOC108348137
|
armadillo repeat-containing X-linked protein 1 |
| chr2_+_187218851 | 0.04 |
ENSRNOT00000017798
|
Sh2d2a
|
SH2 domain containing 2A |
| chr1_+_189432604 | 0.04 |
ENSRNOT00000034610
|
Acsm4
|
acyl-CoA synthetase medium-chain family member 4 |
| chr3_+_112242270 | 0.04 |
ENSRNOT00000080533
ENSRNOT00000082876 |
Capn3
|
calpain 3 |
| chr1_-_224994653 | 0.04 |
ENSRNOT00000026313
|
Polr2g
|
RNA polymerase II subunit G |
| chr5_-_144345531 | 0.04 |
ENSRNOT00000014721
|
Tekt2
|
tektin 2 |
| chr2_-_140464607 | 0.04 |
ENSRNOT00000058190
|
Ndufc1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr16_+_72268943 | 0.04 |
ENSRNOT00000032116
|
Ido2
|
indoleamine 2,3-dioxygenase 2 |
| chr14_+_81043454 | 0.04 |
ENSRNOT00000043609
|
AC114393.1
|
|
| chr6_+_108796182 | 0.04 |
ENSRNOT00000006297
|
Fcf1
|
FCF1 rRNA-processing protein |
| chr4_+_147832136 | 0.04 |
ENSRNOT00000064603
|
Rho
|
rhodopsin |
| chr1_-_67284864 | 0.04 |
ENSRNOT00000082908
|
LOC691661
|
similar to zinc finger and SCAN domain containing 4 |
| chr16_-_45929 | 0.04 |
ENSRNOT00000043153
|
AABR07024439.1
|
|
| chr8_-_107490093 | 0.04 |
ENSRNOT00000046832
|
LOC684466
|
similar to Fas apoptotic inhibitory molecule 1 (rFAIM) |
| chr9_-_53315915 | 0.04 |
ENSRNOT00000038093
|
Mstn
|
myostatin |
| chr1_-_198839228 | 0.04 |
ENSRNOT00000052100
|
LOC102553866
|
zinc finger protein 764-like |
| chr15_-_42794279 | 0.04 |
ENSRNOT00000023385
|
Ephx2
|
epoxide hydrolase 2 |
| chrX_+_31984612 | 0.04 |
ENSRNOT00000005181
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chr3_+_170994038 | 0.04 |
ENSRNOT00000081823
|
Spo11
|
SPO11, initiator of meiotic double stranded breaks |
| chrX_+_15225645 | 0.04 |
ENSRNOT00000008648
|
Glod5
|
glyoxalase domain containing 5 |
| chr14_+_84231639 | 0.04 |
ENSRNOT00000066362
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr9_+_72052966 | 0.04 |
ENSRNOT00000021099
|
Pth2r
|
parathyroid hormone 2 receptor |
| chrX_+_123751293 | 0.04 |
ENSRNOT00000089883
|
Nkap
|
NFKB activating protein |
| chr4_+_9160067 | 0.04 |
ENSRNOT00000090600
ENSRNOT00000015426 |
Orc5
|
origin recognition complex, subunit 5 |
| chr10_-_14072230 | 0.04 |
ENSRNOT00000018405
|
Tbl3
|
transducin (beta)-like 3 |
| chr4_+_151298548 | 0.04 |
ENSRNOT00000010746
|
Cacna2d4
|
calcium voltage-gated channel auxiliary subunit alpha2delta 4 |
| chr10_+_90984227 | 0.04 |
ENSRNOT00000003890
ENSRNOT00000093707 |
Ccdc103
|
coiled-coil domain containing 103 |
| chr5_+_61425746 | 0.04 |
ENSRNOT00000064113
|
RGD1305807
|
hypothetical LOC298077 |
| chr1_-_237910012 | 0.04 |
ENSRNOT00000023664
|
Anxa1
|
annexin A1 |
| chr20_+_13817795 | 0.04 |
ENSRNOT00000036518
|
Gstt3
|
glutathione S-transferase, theta 3 |
| chr13_+_60545635 | 0.04 |
ENSRNOT00000077153
|
Uchl5
|
ubiquitin C-terminal hydrolase L5 |
| chr9_-_10441834 | 0.04 |
ENSRNOT00000043704
|
Rpl36
|
ribosomal protein L36 |
| chr5_+_6373583 | 0.04 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr11_+_86715981 | 0.04 |
ENSRNOT00000050269
|
Comt
|
catechol-O-methyltransferase |
| chr5_+_35865605 | 0.04 |
ENSRNOT00000011442
|
Ccnc
|
cyclin C |
| chr12_+_38965063 | 0.04 |
ENSRNOT00000074936
|
Morn3
|
MORN repeat containing 3 |
| chr8_-_116391158 | 0.04 |
ENSRNOT00000078720
ENSRNOT00000022550 |
Gnai2
|
G protein subunit alpha i2 |
| chr3_+_103747654 | 0.04 |
ENSRNOT00000006887
|
Nop10
|
NOP10 ribonucleoprotein |
| chr8_-_85645718 | 0.04 |
ENSRNOT00000032185
|
Gsta2
|
glutathione S-transferase alpha 2 |
| chr13_+_93939022 | 0.04 |
ENSRNOT00000088204
|
Exo1
|
exonuclease 1 |
| chr12_+_19335841 | 0.04 |
ENSRNOT00000001831
|
Mblac1
|
metallo-beta-lactamase domain containing 1 |
| chr1_-_199341302 | 0.04 |
ENSRNOT00000073596
|
Vkorc1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr10_+_34402482 | 0.04 |
ENSRNOT00000072317
|
Tpcr12
|
putative olfactory receptor |
| chr10_+_85257876 | 0.04 |
ENSRNOT00000014752
|
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chr16_+_54899452 | 0.03 |
ENSRNOT00000016882
|
Cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr7_-_49741540 | 0.03 |
ENSRNOT00000006523
|
Myf6
|
myogenic factor 6 |
| chr5_-_133067245 | 0.03 |
ENSRNOT00000033163
|
Skint10
|
selection and upkeep of intraepithelial T cells 10 |
| chr15_+_48674380 | 0.03 |
ENSRNOT00000018762
|
Fbxo16
|
F-box protein 16 |
| chr3_-_133232322 | 0.03 |
ENSRNOT00000034487
|
Esf1
|
ESF1 nucleolar pre-rRNA processing protein homolog |
| chr10_-_105228547 | 0.03 |
ENSRNOT00000028875
|
Srp68
|
signal recognition particle 68 |
| chr10_-_14613878 | 0.03 |
ENSRNOT00000024072
|
Baiap3
|
BAI1-associated protein 3 |
| chr5_+_126668689 | 0.03 |
ENSRNOT00000036072
|
Cyb5rl
|
cytochrome b5 reductase-like |
| chr3_-_8659102 | 0.03 |
ENSRNOT00000050908
|
Zdhhc12
|
zinc finger, DHHC-type containing 12 |
| chr12_-_19440501 | 0.03 |
ENSRNOT00000060048
|
Nxpe5
|
neurexophilin and PC-esterase domain family, member 5 |
| chr2_-_27949066 | 0.03 |
ENSRNOT00000081472
ENSRNOT00000022138 |
Nsa2
|
NSA2 ribosome biogenesis homolog |
| chr5_+_33097654 | 0.03 |
ENSRNOT00000008087
|
Cngb3
|
cyclic nucleotide gated channel beta 3 |
| chr1_+_5448958 | 0.03 |
ENSRNOT00000061930
|
Epm2a
|
epilepsy, progressive myoclonus type 2A |
| chr10_+_15495684 | 0.03 |
ENSRNOT00000027662
|
Mrpl28
|
mitochondrial ribosomal protein L28 |
| chr9_+_88918433 | 0.03 |
ENSRNOT00000021730
|
Ccl20
|
C-C motif chemokine ligand 20 |
| chr9_+_90857308 | 0.03 |
ENSRNOT00000073993
|
LOC100911572
|
collagen alpha-3(IV) chain-like |
| chrX_-_64908682 | 0.03 |
ENSRNOT00000084107
|
Zc4h2
|
zinc finger C4H2-type containing |
| chr19_+_30936703 | 0.03 |
ENSRNOT00000024568
|
Smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr18_+_70974885 | 0.03 |
ENSRNOT00000025198
|
RGD1562987
|
similar to cDNA sequence BC031181 |
| chr12_-_41448668 | 0.03 |
ENSRNOT00000001856
|
Rasal1
|
RAS protein activator like 1 (GAP1 like) |
| chrX_+_124894706 | 0.03 |
ENSRNOT00000066677
|
Mcts1
|
MCTS1, re-initiation and release factor |
| chr8_-_119326938 | 0.03 |
ENSRNOT00000044467
|
Ccrl2
|
C-C motif chemokine receptor like 2 |
| chr3_+_173969273 | 0.03 |
ENSRNOT00000083970
|
Cdh26
|
cadherin 26 |
| chr1_-_101741441 | 0.03 |
ENSRNOT00000028570
|
Sult2b1
|
sulfotransferase family 2B member 1 |
| chr2_-_259382765 | 0.03 |
ENSRNOT00000091407
|
St6galnac3
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr16_+_37177443 | 0.03 |
ENSRNOT00000014083
|
Cep44
|
centrosomal protein 44 |
| chr2_-_256915563 | 0.03 |
ENSRNOT00000084873
ENSRNOT00000029990 |
Ifi44
|
interferon-induced protein 44 |
| chr1_-_229601032 | 0.03 |
ENSRNOT00000016690
|
Cntf
|
ciliary neurotrophic factor |
| chr1_-_81627710 | 0.03 |
ENSRNOT00000071704
|
LOC100909700
|
CD177 antigen-like |
| chr3_+_11424099 | 0.03 |
ENSRNOT00000019184
|
Ptges2
|
prostaglandin E synthase 2 |
| chrX_-_79909678 | 0.03 |
ENSRNOT00000050336
|
Brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr13_-_79890134 | 0.03 |
ENSRNOT00000083043
|
RGD1309106
|
similar to hypothetical protein |
| chr9_-_10182676 | 0.03 |
ENSRNOT00000074894
|
LOC316124
|
similar to gonadotropin-regulated long chain acyl-CoA synthetase |
| chr1_-_196883051 | 0.03 |
ENSRNOT00000055034
|
Nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr16_-_68913628 | 0.03 |
ENSRNOT00000016656
|
Chrnb3
|
cholinergic receptor nicotinic beta 3 subunit |
| chr7_-_116063078 | 0.03 |
ENSRNOT00000076932
ENSRNOT00000035496 |
Gml
|
glycosylphosphatidylinositol anchored molecule like |
| chrX_-_25628272 | 0.03 |
ENSRNOT00000086414
|
Mid1
|
midline 1 |
| chr10_+_13797562 | 0.03 |
ENSRNOT00000011784
ENSRNOT00000083312 |
Eci1
|
enoyl-CoA delta isomerase 1 |
| chr1_+_85213652 | 0.03 |
ENSRNOT00000092044
|
Nccrp1
|
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
| chr1_-_100980340 | 0.03 |
ENSRNOT00000064272
|
Prmt1
|
protein arginine methyltransferase 1 |
| chr3_-_137969658 | 0.03 |
ENSRNOT00000007727
|
Bfsp1
|
beaded filament structural protein 1 |
| chr9_-_93125014 | 0.03 |
ENSRNOT00000023829
|
Htr2b
|
5-hydroxytryptamine receptor 2B |
| chr4_+_95498003 | 0.03 |
ENSRNOT00000008358
|
Atoh1
|
atonal bHLH transcription factor 1 |
| chr9_+_51298426 | 0.03 |
ENSRNOT00000065212
|
Gulp1
|
GULP, engulfment adaptor PTB domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Crx_Gsc
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071663 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) |
| 0.0 | 0.1 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.2 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.0 | 0.1 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.0 | 0.1 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.0 | 0.1 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.1 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.0 | 0.0 | GO:0006532 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.0 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.1 | GO:1902569 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.0 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.0 | 0.0 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 0.0 | 0.0 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.0 | GO:0046864 | isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.0 | 0.0 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.1 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.0 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.0 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:1990005 | granular vesicle(GO:1990005) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.2 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.0 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.0 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.0 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.0 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.0 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |