Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Creb3l2
Z-value: 0.25
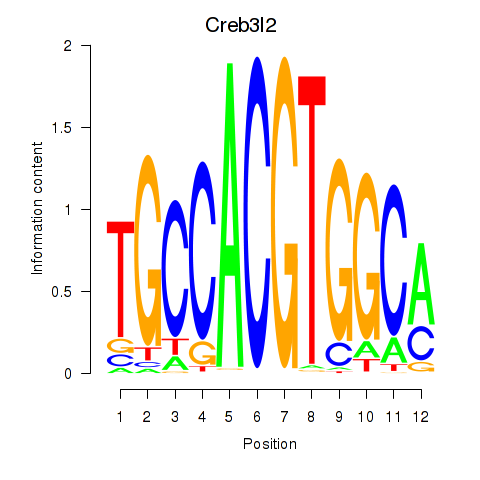
Transcription factors associated with Creb3l2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Creb3l2
|
ENSRNOG00000012826 | cAMP responsive element binding protein 3-like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Creb3l2 | rn6_v1_chr4_-_64981384_64981384 | -0.49 | 4.0e-01 | Click! |
Activity profile of Creb3l2 motif
Sorted Z-values of Creb3l2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_15098904 | 0.08 |
ENSRNOT00000007367
ENSRNOT00000087033 |
Rbm3
|
RNA binding motif (RNP1, RRM) protein 3 |
| chr7_+_141249044 | 0.06 |
ENSRNOT00000084911
|
Aqp5
|
aquaporin 5 |
| chr11_-_70499200 | 0.05 |
ENSRNOT00000002439
|
Slc12a8
|
solute carrier family 12, member 8 |
| chr7_+_3332788 | 0.05 |
ENSRNOT00000010180
|
Cd63
|
Cd63 molecule |
| chr3_-_10375826 | 0.05 |
ENSRNOT00000093365
|
Ass1
|
argininosuccinate synthase 1 |
| chr7_+_69213147 | 0.04 |
ENSRNOT00000032786
|
Lrig3
|
leucine-rich repeats and immunoglobulin-like domains 3 |
| chr10_-_16752205 | 0.04 |
ENSRNOT00000079462
|
Crebrf
|
CREB3 regulatory factor |
| chr3_+_55094637 | 0.04 |
ENSRNOT00000058763
|
Cers6
|
ceramide synthase 6 |
| chr5_-_166726794 | 0.04 |
ENSRNOT00000022799
|
Slc25a33
|
solute carrier family 25 member 33 |
| chr7_+_53878610 | 0.04 |
ENSRNOT00000091910
|
Osbpl8
|
oxysterol binding protein-like 8 |
| chr5_-_126221916 | 0.04 |
ENSRNOT00000052130
|
Lexm
|
lymphocyte expansion molecule |
| chr5_-_152458023 | 0.04 |
ENSRNOT00000031225
|
Cnksr1
|
connector enhancer of kinase suppressor of Ras 1 |
| chr7_-_124198200 | 0.04 |
ENSRNOT00000078947
|
Arfgap3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr1_+_15620653 | 0.03 |
ENSRNOT00000017401
|
Map7
|
microtubule-associated protein 7 |
| chr19_-_37210412 | 0.03 |
ENSRNOT00000083097
|
B3gnt9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
| chrX_-_10413984 | 0.03 |
ENSRNOT00000039551
ENSRNOT00000091448 |
Ddx3x
|
DEAD-box helicase 3, X-linked |
| chr7_-_24667301 | 0.03 |
ENSRNOT00000009154
|
Tmem263
|
transmembrane protein 263 |
| chr4_-_34779776 | 0.03 |
ENSRNOT00000077329
|
Ica1
|
islet cell autoantigen 1 |
| chr17_+_85356042 | 0.03 |
ENSRNOT00000022201
|
Commd3
|
COMM domain containing 3 |
| chr5_+_135574172 | 0.03 |
ENSRNOT00000023416
|
Tesk2
|
testis-specific kinase 2 |
| chr9_+_94425252 | 0.03 |
ENSRNOT00000064965
ENSRNOT00000076099 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr10_-_37209881 | 0.03 |
ENSRNOT00000090475
|
Sec24a
|
SEC24 homolog A, COPII coat complex component |
| chr11_-_83867203 | 0.03 |
ENSRNOT00000002394
|
Chrd
|
chordin |
| chr8_-_58253688 | 0.03 |
ENSRNOT00000010956
|
Cul5
|
cullin 5 |
| chr1_-_229639187 | 0.03 |
ENSRNOT00000016804
|
Zfp91
|
zinc finger protein 91 |
| chr18_-_33414236 | 0.03 |
ENSRNOT00000020385
|
Yipf5
|
Yip1 domain family, member 5 |
| chr3_-_66417741 | 0.03 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr4_-_34780193 | 0.02 |
ENSRNOT00000011651
|
Ica1
|
islet cell autoantigen 1 |
| chr20_+_6288267 | 0.02 |
ENSRNOT00000000627
|
Srsf3
|
serine and arginine rich splicing factor 3 |
| chr6_+_42852683 | 0.02 |
ENSRNOT00000079185
|
Odc1
|
ornithine decarboxylase 1 |
| chr3_+_114176309 | 0.02 |
ENSRNOT00000023350
|
Sord
|
sorbitol dehydrogenase |
| chr9_+_69953440 | 0.02 |
ENSRNOT00000034740
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr6_+_108167716 | 0.02 |
ENSRNOT00000064426
|
Lin52
|
lin-52 DREAM MuvB core complex component |
| chr20_-_33521118 | 0.02 |
ENSRNOT00000083295
|
AABR07045071.1
|
|
| chr3_-_81282157 | 0.02 |
ENSRNOT00000051258
|
Large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr14_+_104475082 | 0.02 |
ENSRNOT00000084481
|
Rab1a
|
RAB1A, member RAS oncogene family |
| chr1_+_162676269 | 0.02 |
ENSRNOT00000017702
|
Clns1a
|
chloride nucleotide-sensitive channel 1A |
| chr4_+_113935492 | 0.02 |
ENSRNOT00000035329
|
LOC103692170
|
coiled-coil domain-containing protein 142 |
| chr5_-_138470096 | 0.02 |
ENSRNOT00000011392
|
Ppcs
|
phosphopantothenoylcysteine synthetase |
| chr17_+_9837402 | 0.02 |
ENSRNOT00000076436
|
Rab24
|
RAB24, member RAS oncogene family |
| chr1_+_266333440 | 0.02 |
ENSRNOT00000071243
|
Sfxn2
|
sideroflexin 2 |
| chr17_-_21591203 | 0.02 |
ENSRNOT00000036195
|
Pak1ip1
|
PAK1 interacting protein 1 |
| chr4_-_87257667 | 0.02 |
ENSRNOT00000083491
|
Nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr9_+_99998275 | 0.02 |
ENSRNOT00000074395
|
Gpc1
|
glypican 1 |
| chr2_-_204032023 | 0.02 |
ENSRNOT00000040430
|
Atp1a1
|
ATPase Na+/K+ transporting subunit alpha 1 |
| chr1_+_215701544 | 0.02 |
ENSRNOT00000027585
|
Mrpl23
|
mitochondrial ribosomal protein L23 |
| chr5_+_154668179 | 0.02 |
ENSRNOT00000015971
|
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr10_+_70134729 | 0.02 |
ENSRNOT00000076739
|
Lig3
|
DNA ligase 3 |
| chr4_-_58006839 | 0.02 |
ENSRNOT00000076645
|
Cep41
|
centrosomal protein 41 |
| chr15_-_60766579 | 0.02 |
ENSRNOT00000079978
|
Akap11
|
A-kinase anchoring protein 11 |
| chr5_+_60250546 | 0.02 |
ENSRNOT00000017707
|
Zcchc7
|
zinc finger CCHC-type containing 7 |
| chr4_+_98648545 | 0.01 |
ENSRNOT00000008451
|
Eif2ak3
|
eukaryotic translation initiation factor 2 alpha kinase 3 |
| chr12_+_16912249 | 0.01 |
ENSRNOT00000085936
|
Tmem184a
|
transmembrane protein 184A |
| chr2_-_53827175 | 0.01 |
ENSRNOT00000078158
|
RGD1305938
|
similar to expressed sequence AW549877 |
| chr2_+_189400696 | 0.01 |
ENSRNOT00000046919
ENSRNOT00000089801 |
LOC361985
|
similar to NICE-3 |
| chr3_+_122803772 | 0.01 |
ENSRNOT00000009564
|
Nop56
|
NOP56 ribonucleoprotein |
| chr10_+_34938667 | 0.01 |
ENSRNOT00000086443
ENSRNOT00000059104 |
LOC103690164
|
5-phosphohydroxy-L-lysine phospho-lyase |
| chr2_-_211017778 | 0.01 |
ENSRNOT00000026883
|
Sypl2
|
synaptophysin-like 2 |
| chr1_-_141893674 | 0.01 |
ENSRNOT00000019059
ENSRNOT00000085988 |
Idh2
|
isocitrate dehydrogenase (NADP(+)) 2, mitochondrial |
| chr18_-_15637715 | 0.01 |
ENSRNOT00000022270
|
Dsg2
|
desmoglein 2 |
| chr1_-_222098297 | 0.01 |
ENSRNOT00000028678
|
Rps6ka4
|
ribosomal protein S6 kinase A4 |
| chr19_-_49448072 | 0.01 |
ENSRNOT00000014997
|
Cmc2
|
C-x(9)-C motif containing 2 |
| chr3_-_119405300 | 0.01 |
ENSRNOT00000015568
|
Sppl2a
|
signal peptide peptidase-like 2A |
| chr2_+_44512895 | 0.01 |
ENSRNOT00000013218
|
Slc38a9
|
solute carrier family 38, member 9 |
| chr12_+_24473981 | 0.01 |
ENSRNOT00000001973
|
Fzd9
|
frizzled class receptor 9 |
| chr5_+_138470069 | 0.01 |
ENSRNOT00000076343
ENSRNOT00000064073 |
Zmynd12
|
zinc finger, MYND-type containing 12 |
| chr1_+_170471238 | 0.01 |
ENSRNOT00000076961
ENSRNOT00000075597 ENSRNOT00000076631 ENSRNOT00000076783 |
Timm10b
Dnhd1
|
translocase of inner mitochondrial membrane 10B dynein heavy chain domain 1 |
| chr15_-_34322115 | 0.01 |
ENSRNOT00000026734
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr10_+_36099263 | 0.01 |
ENSRNOT00000083568
|
Adamts2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr10_-_88667345 | 0.01 |
ENSRNOT00000025518
|
Kcnh4
|
potassium voltage-gated channel subfamily H member 4 |
| chr10_+_56610051 | 0.01 |
ENSRNOT00000024348
|
Dvl2
|
dishevelled segment polarity protein 2 |
| chr3_+_85544827 | 0.01 |
ENSRNOT00000051901
|
AABR07052953.1
|
|
| chr19_+_10827371 | 0.01 |
ENSRNOT00000024120
|
Fam192a
|
family with sequence similarity 192, member A |
| chrX_-_154722220 | 0.01 |
ENSRNOT00000089743
ENSRNOT00000087893 |
Fmr1
|
fragile X mental retardation 1 |
| chr19_-_57422093 | 0.01 |
ENSRNOT00000025512
|
RGD1559896
|
similar to RIKEN cDNA 2310022B05 |
| chr13_+_105408179 | 0.01 |
ENSRNOT00000003378
|
Ddx3y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chrX_+_16050780 | 0.01 |
ENSRNOT00000079054
|
Clcn5
|
chloride voltage-gated channel 5 |
| chr6_+_26797126 | 0.01 |
ENSRNOT00000010586
|
Cgref1
|
cell growth regulator with EF hand domain 1 |
| chr19_-_38120578 | 0.01 |
ENSRNOT00000026873
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr7_+_72924799 | 0.01 |
ENSRNOT00000008969
|
Laptm4b
|
lysosomal protein transmembrane 4 beta |
| chr7_-_73270308 | 0.01 |
ENSRNOT00000007430
|
Rida
|
reactive intermediate imine deaminase A homolog |
| chr6_-_137733026 | 0.01 |
ENSRNOT00000019213
|
Jag2
|
jagged 2 |
| chr2_+_250600823 | 0.01 |
ENSRNOT00000083750
|
Sep15
|
selenoprotein 15 |
| chr3_-_119405453 | 0.01 |
ENSRNOT00000090355
|
Sppl2a
|
signal peptide peptidase-like 2A |
| chrX_+_138046494 | 0.01 |
ENSRNOT00000010596
|
Stk26
|
serine/threonine kinase 26 |
| chr11_-_16889201 | 0.01 |
ENSRNOT00000002121
|
Btg3
|
BTG anti-proliferation factor 3 |
| chr7_-_143738237 | 0.01 |
ENSRNOT00000055320
|
Spryd3
|
SPRY domain containing 3 |
| chr20_-_10386663 | 0.01 |
ENSRNOT00000045275
ENSRNOT00000042432 |
Cbs
|
cystathionine beta synthase |
| chr19_-_43215281 | 0.01 |
ENSRNOT00000025052
|
Aars
|
alanyl-tRNA synthetase |
| chr2_+_209097927 | 0.01 |
ENSRNOT00000023807
|
Dennd2d
|
DENN domain containing 2D |
| chr3_-_95418679 | 0.01 |
ENSRNOT00000018074
|
Rcn1
|
reticulocalbin 1 |
| chr5_-_173081839 | 0.00 |
ENSRNOT00000066880
|
Mmp23
|
matrix metallopeptidase 23 |
| chr1_+_48176106 | 0.00 |
ENSRNOT00000021840
|
Igf2r
|
insulin-like growth factor 2 receptor |
| chr10_-_104564480 | 0.00 |
ENSRNOT00000008525
|
Galk1
|
galactokinase 1 |
| chr20_+_22728208 | 0.00 |
ENSRNOT00000000794
|
LOC100363472
|
nuclear receptor-binding factor 2-like |
| chr15_+_45422010 | 0.00 |
ENSRNOT00000012231
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chr1_+_154377447 | 0.00 |
ENSRNOT00000084268
ENSRNOT00000092086 ENSRNOT00000091470 ENSRNOT00000025415 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr16_-_21348391 | 0.00 |
ENSRNOT00000083537
|
Lpar2
|
lysophosphatidic acid receptor 2 |
| chr4_-_49439867 | 0.00 |
ENSRNOT00000090583
|
Fam3c
|
family with sequence similarity 3, member C |
| chr8_-_98738446 | 0.00 |
ENSRNOT00000019860
|
Zic1
|
Zic family member 1 |
| chr3_-_175709465 | 0.00 |
ENSRNOT00000089971
|
Gata5
|
GATA binding protein 5 |
| chrX_+_68627313 | 0.00 |
ENSRNOT00000076699
ENSRNOT00000076795 ENSRNOT00000008705 |
Yipf6
|
Yip1 domain family, member 6 |
| chr19_-_43215077 | 0.00 |
ENSRNOT00000082151
|
Aars
|
alanyl-tRNA synthetase |
| chr2_+_189106039 | 0.00 |
ENSRNOT00000028210
|
Ube2q1
|
ubiquitin conjugating enzyme E2 Q1 |
| chr9_-_61690956 | 0.00 |
ENSRNOT00000066589
|
Hspd1
|
heat shock protein family D member 1 |
| chr10_+_82375572 | 0.00 |
ENSRNOT00000004947
|
Mrpl27
|
mitochondrial ribosomal protein L27 |
| chr4_+_119815139 | 0.00 |
ENSRNOT00000083402
ENSRNOT00000016137 |
Hmces
|
5-hydroxymethylcytosine (hmC) binding, ES cell-specific |
| chr14_+_35683442 | 0.00 |
ENSRNOT00000003085
|
Chic2
|
cysteine-rich hydrophobic domain 2 |
| chr1_-_82120902 | 0.00 |
ENSRNOT00000027684
|
Erf
|
Ets2 repressor factor |
| chr10_+_88326080 | 0.00 |
ENSRNOT00000076475
|
Fkbp10
|
FK506 binding protein 10 |
| chr15_+_38096994 | 0.00 |
ENSRNOT00000064635
|
Mrpl57
|
mitochondrial ribosomal protein L57 |
| chr20_-_30888735 | 0.00 |
ENSRNOT00000090004
ENSRNOT00000063875 |
Adamts14
|
ADAM metallopeptidase with thrombospondin type 1 motif, 14 |
| chr6_-_55524436 | 0.00 |
ENSRNOT00000006752
|
Tspan13
|
tetraspanin 13 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Creb3l2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0000053 | argininosuccinate metabolic process(GO:0000053) |
| 0.0 | 0.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.0 | GO:1902211 | regulation of prolactin signaling pathway(GO:1902211) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |