Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
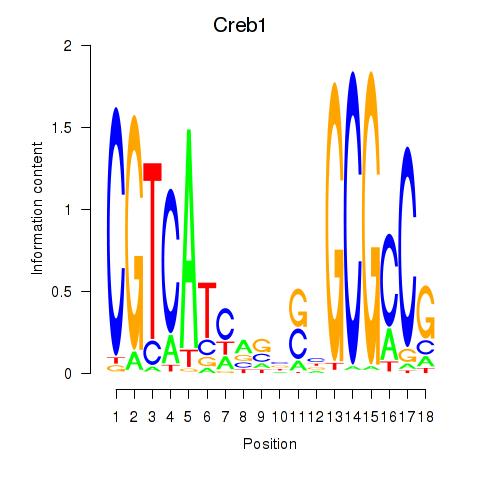
Results for Creb1
Z-value: 0.76
Transcription factors associated with Creb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Creb1
|
ENSRNOG00000013412 | cAMP responsive element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Creb1 | rn6_v1_chr9_+_71230108_71230108 | 0.57 | 3.2e-01 | Click! |
Activity profile of Creb1 motif
Sorted Z-values of Creb1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_219412816 | 0.44 |
ENSRNOT00000083204
ENSRNOT00000029580 |
Rps6kb2
|
ribosomal protein S6 kinase B2 |
| chr6_+_144156175 | 0.40 |
ENSRNOT00000006582
|
Esyt2
|
extended synaptotagmin 2 |
| chr10_-_104482838 | 0.35 |
ENSRNOT00000007246
|
Recql5
|
RecQ like helicase 5 |
| chr17_+_18358712 | 0.30 |
ENSRNOT00000001979
|
Nup153
|
nucleoporin 153 |
| chr8_-_49271834 | 0.27 |
ENSRNOT00000085022
|
Ube4a
|
ubiquitination factor E4A |
| chr1_-_198450688 | 0.26 |
ENSRNOT00000027392
|
Pagr1
|
Paxip1-associated glutamate-rich protein 1 |
| chr10_-_110555629 | 0.25 |
ENSRNOT00000054918
ENSRNOT00000088474 |
Wdr45b
|
WD repeat domain 45B |
| chr20_+_6205903 | 0.25 |
ENSRNOT00000092333
ENSRNOT00000092655 |
Kctd20
|
potassium channel tetramerization domain containing 20 |
| chr20_+_4967194 | 0.23 |
ENSRNOT00000070846
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr11_+_71796282 | 0.21 |
ENSRNOT00000087489
|
Fbxo45
|
F-box protein 45 |
| chr1_+_221710670 | 0.21 |
ENSRNOT00000064798
|
Map4k2
|
mitogen activated protein kinase kinase kinase kinase 2 |
| chr7_+_11033317 | 0.20 |
ENSRNOT00000007498
|
Gna11
|
guanine nucleotide binding protein, alpha 11 |
| chr1_-_175657485 | 0.20 |
ENSRNOT00000024561
|
rnf141
|
ring finger protein 141 |
| chr15_+_34520142 | 0.20 |
ENSRNOT00000074659
|
Nynrin
|
NYN domain and retroviral integrase containing |
| chr18_+_1142782 | 0.19 |
ENSRNOT00000046718
|
Thoc1
|
THO complex 1 |
| chr13_+_99136871 | 0.19 |
ENSRNOT00000078263
ENSRNOT00000004350 |
Sde2
|
SDE2 telomere maintenance homolog |
| chr14_+_1469748 | 0.18 |
ENSRNOT00000071648
|
Ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr3_-_176791960 | 0.17 |
ENSRNOT00000018237
|
Gmeb2
|
glucocorticoid modulatory element binding protein 2 |
| chr6_-_122545899 | 0.17 |
ENSRNOT00000005175
|
Kcnk10
|
potassium two pore domain channel subfamily K member 10 |
| chr6_+_137824213 | 0.17 |
ENSRNOT00000056880
|
Pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr13_-_109629482 | 0.16 |
ENSRNOT00000072452
|
Flvcr1
|
feline leukemia virus subgroup C cellular receptor 1 |
| chr7_+_144647587 | 0.16 |
ENSRNOT00000022398
|
Hoxc4
|
homeo box C4 |
| chr1_-_182140570 | 0.16 |
ENSRNOT00000060098
|
Ythdc2
|
YTH domain containing 2 |
| chr11_-_82664554 | 0.15 |
ENSRNOT00000002425
ENSRNOT00000087105 |
Senp2
|
Sumo1/sentrin/SMT3 specific peptidase 2 |
| chr8_+_69127708 | 0.15 |
ENSRNOT00000013490
|
Snapc5
|
small nuclear RNA activating complex, polypeptide 5 |
| chr18_-_63357194 | 0.15 |
ENSRNOT00000089408
ENSRNOT00000066103 |
Spire1
|
spire-type actin nucleation factor 1 |
| chr11_+_61748883 | 0.15 |
ENSRNOT00000093552
|
Qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr6_-_122897997 | 0.15 |
ENSRNOT00000057601
|
Eml5
|
echinoderm microtubule associated protein like 5 |
| chr1_-_190370499 | 0.15 |
ENSRNOT00000084389
|
AABR07005618.1
|
|
| chr2_-_28370261 | 0.15 |
ENSRNOT00000022259
|
Utp15
|
UTP15, small subunit processome component |
| chr9_+_16647598 | 0.15 |
ENSRNOT00000087413
|
Klc4
|
kinesin light chain 4 |
| chr20_-_7654428 | 0.15 |
ENSRNOT00000045044
|
Tcp11
|
t-complex 11 |
| chr2_+_205553163 | 0.14 |
ENSRNOT00000039572
|
Nras
|
neuroblastoma RAS viral oncogene homolog |
| chr1_+_212230259 | 0.14 |
ENSRNOT00000034426
|
Kndc1
|
kinase non-catalytic C-lobe domain containing 1 |
| chr1_+_81395841 | 0.14 |
ENSRNOT00000072750
|
Irgq
|
immunity-related GTPase Q |
| chr12_-_52072106 | 0.14 |
ENSRNOT00000088306
ENSRNOT00000068621 |
Ddx51
|
DEAD-box helicase 51 |
| chr1_+_56982821 | 0.13 |
ENSRNOT00000059845
|
Ermard
|
ER membrane-associated RNA degradation |
| chr8_+_114866768 | 0.13 |
ENSRNOT00000076731
|
Wdr82
|
WD repeat domain 82 |
| chr9_-_82410904 | 0.13 |
ENSRNOT00000026081
|
Glb1l
|
galactosidase, beta 1-like |
| chr8_+_132032944 | 0.13 |
ENSRNOT00000089278
|
Kif15
|
kinesin family member 15 |
| chr20_-_27308069 | 0.13 |
ENSRNOT00000056047
|
Slc25a16
|
solute carrier family 25 member 16 |
| chr20_+_28027054 | 0.13 |
ENSRNOT00000071386
ENSRNOT00000001044 |
Ranbp2
|
RAN binding protein 2 |
| chr10_-_55589978 | 0.13 |
ENSRNOT00000007087
|
LOC100912917
|
phosphoribosylformylglycinamidine synthase-like |
| chr11_-_86890390 | 0.13 |
ENSRNOT00000046183
|
Trmt2a
|
tRNA methyltransferase 2 homolog A |
| chr6_+_110624856 | 0.13 |
ENSRNOT00000014017
|
Vash1
|
vasohibin 1 |
| chr3_+_110835683 | 0.12 |
ENSRNOT00000072130
|
LOC100911166
|
RNA pseudouridylate synthase domain-containing protein 2-like |
| chr6_+_96171756 | 0.12 |
ENSRNOT00000029328
|
Slc38a6
|
solute carrier family 38, member 6 |
| chr1_+_266143818 | 0.12 |
ENSRNOT00000026987
|
Sufu
|
SUFU negative regulator of hedgehog signaling |
| chr1_-_103128743 | 0.12 |
ENSRNOT00000075006
|
Spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr14_-_80355420 | 0.12 |
ENSRNOT00000049798
|
Acox3
|
acyl-CoA oxidase 3, pristanoyl |
| chr13_+_44475970 | 0.11 |
ENSRNOT00000024602
ENSRNOT00000091645 |
Ccnt2
|
cyclin T2 |
| chr12_-_2170504 | 0.11 |
ENSRNOT00000001309
|
Xab2
|
XPA binding protein 2 |
| chr7_+_130326600 | 0.11 |
ENSRNOT00000084776
ENSRNOT00000055808 |
Ncaph2
|
non-SMC condensin II complex, subunit H2 |
| chr9_+_50664150 | 0.11 |
ENSRNOT00000015393
|
Tpp2
|
tripeptidyl peptidase 2 |
| chr1_-_242861767 | 0.11 |
ENSRNOT00000021185
|
Cbwd1
|
COBW domain containing 1 |
| chr4_+_100209951 | 0.11 |
ENSRNOT00000015807
|
LOC691113
|
hypothetical protein LOC691113 |
| chr18_-_77535564 | 0.11 |
ENSRNOT00000041380
ENSRNOT00000090265 |
Atp9b
|
ATPase phospholipid transporting 9B (putative) |
| chr19_-_56731372 | 0.11 |
ENSRNOT00000024182
|
Nup133
|
nucleoporin 133 |
| chr15_-_24056073 | 0.11 |
ENSRNOT00000015100
|
Wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr1_-_89042176 | 0.10 |
ENSRNOT00000080842
|
Kmt2b
|
lysine methyltransferase 2B |
| chr2_+_56426367 | 0.10 |
ENSRNOT00000016036
|
Lifr
|
leukemia inhibitory factor receptor alpha |
| chr20_-_7482747 | 0.10 |
ENSRNOT00000038195
|
Taf11
|
TATA-box binding protein associated factor 11 |
| chr1_-_103014388 | 0.10 |
ENSRNOT00000045719
|
Uevld
|
UEV and lactate/malate dehyrogenase domains |
| chr2_-_27364906 | 0.10 |
ENSRNOT00000078639
|
Polk
|
DNA polymerase kappa |
| chr11_+_87522971 | 0.10 |
ENSRNOT00000043545
|
Smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr10_+_64398339 | 0.10 |
ENSRNOT00000056278
|
Mrm3
|
mitochondrial rRNA methyltransferase 3 |
| chr5_+_124690214 | 0.10 |
ENSRNOT00000011237
|
Plpp3
|
phospholipid phosphatase 3 |
| chr19_-_54722563 | 0.10 |
ENSRNOT00000025784
|
Slc7a5
|
solute carrier family 7 member 5 |
| chr12_+_37444072 | 0.10 |
ENSRNOT00000077380
ENSRNOT00000092699 ENSRNOT00000001373 ENSRNOT00000092416 |
Eif2b1
|
eukaryotic translation initiation factor 2B subunit 1 alpha |
| chr10_+_81693770 | 0.09 |
ENSRNOT00000003777
ENSRNOT00000085681 |
Spag9
|
sperm associated antigen 9 |
| chr1_-_89124132 | 0.09 |
ENSRNOT00000031044
|
Haus5
|
HAUS augmin-like complex, subunit 5 |
| chr1_+_185356975 | 0.09 |
ENSRNOT00000086681
|
Plekha7
|
pleckstrin homology domain containing A7 |
| chr3_-_111560556 | 0.09 |
ENSRNOT00000030532
|
Ltk
|
leukocyte receptor tyrosine kinase |
| chr19_+_55300395 | 0.09 |
ENSRNOT00000092169
|
Ctu2
|
cytosolic thiouridylase subunit 2 |
| chr9_+_16647922 | 0.09 |
ENSRNOT00000031625
|
Klc4
|
kinesin light chain 4 |
| chr15_-_58171049 | 0.09 |
ENSRNOT00000001371
|
Gpalpp1
|
GPALPP motifs containing 1 |
| chr11_+_89277039 | 0.09 |
ENSRNOT00000002511
|
Mzt2b
|
mitotic spindle organizing protein 2B |
| chr17_+_6665659 | 0.09 |
ENSRNOT00000025980
|
Hnrnpk
|
heterogeneous nuclear ribonucleoprotein K |
| chr14_-_66771750 | 0.09 |
ENSRNOT00000005587
|
Pacrgl
|
PARK2 co-regulated-like |
| chr16_+_32449116 | 0.09 |
ENSRNOT00000050549
|
Clcn3
|
chloride voltage-gated channel 3 |
| chr6_+_26560601 | 0.09 |
ENSRNOT00000077481
|
Gtf3c2
|
general transcription factor IIIC subunit 2 |
| chr16_+_19051965 | 0.09 |
ENSRNOT00000016399
|
Slc35e1
|
solute carrier family 35, member E1 |
| chr1_+_80028928 | 0.08 |
ENSRNOT00000012082
|
Fbxo46
|
F-box protein 46 |
| chr14_+_44580216 | 0.08 |
ENSRNOT00000088674
ENSRNOT00000003907 |
Rfc1
|
replication factor C subunit 1 |
| chr17_+_507377 | 0.08 |
ENSRNOT00000023638
|
Npepo
|
aminopeptidase O |
| chr9_-_28973246 | 0.08 |
ENSRNOT00000091865
ENSRNOT00000015453 |
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr3_+_112519808 | 0.08 |
ENSRNOT00000014129
|
Rpusd2
|
RNA pseudouridylate synthase domain containing 2 |
| chr7_-_137856485 | 0.08 |
ENSRNOT00000007003
|
LOC688906
|
similar to splicing factor, arginine/serine-rich 2, interacting protein |
| chr8_-_36410612 | 0.08 |
ENSRNOT00000091308
|
Foxred1
|
FAD-dependent oxidoreductase domain containing 1 |
| chr19_+_37990374 | 0.08 |
ENSRNOT00000026827
|
Dus2
|
dihydrouridine synthase 2 |
| chr3_+_147713821 | 0.08 |
ENSRNOT00000007558
|
Csnk2a1
|
casein kinase 2 alpha 1 |
| chr19_+_37873515 | 0.07 |
ENSRNOT00000026163
ENSRNOT00000089921 |
Pskh1
|
protein serine kinase H1 |
| chr3_-_140728129 | 0.07 |
ENSRNOT00000015637
|
Ralgapa2
|
Ral GTPase activating protein catalytic alpha subunit 2 |
| chr14_-_1595972 | 0.07 |
ENSRNOT00000075415
|
Pigg
|
phosphatidylinositol glycan anchor biosynthesis, class G |
| chr14_+_17064353 | 0.07 |
ENSRNOT00000003052
|
Scarb2
|
scavenger receptor class B, member 2 |
| chr7_+_108613739 | 0.07 |
ENSRNOT00000007564
|
Phf20l1
|
PHD finger protein 20-like 1 |
| chr7_-_116872511 | 0.07 |
ENSRNOT00000010120
|
Zc3h3
|
zinc finger CCCH type containing 3 |
| chr3_-_9822182 | 0.07 |
ENSRNOT00000076637
ENSRNOT00000010115 |
RGD1305178
|
similar to Hypothetical protein MGC11690 |
| chr19_+_43220508 | 0.07 |
ENSRNOT00000091788
ENSRNOT00000084462 ENSRNOT00000087474 |
Ddx19a
|
DEAD-box helicase 19A |
| chr17_-_13586554 | 0.07 |
ENSRNOT00000041371
|
Secisbp2
|
SECIS binding protein 2 |
| chr4_+_5841998 | 0.07 |
ENSRNOT00000010025
|
Xrcc2
|
X-ray repair cross complementing 2 |
| chr7_+_122636171 | 0.07 |
ENSRNOT00000068666
|
Xpnpep3
|
X-prolyl aminopeptidase 3 |
| chr14_-_2438592 | 0.06 |
ENSRNOT00000072386
|
LOC100910143
|
GPI ethanolamine phosphate transferase 2-like |
| chr5_+_150001281 | 0.06 |
ENSRNOT00000034057
|
Mecr
|
mitochondrial trans-2-enoyl-CoA reductase |
| chr5_+_57291156 | 0.06 |
ENSRNOT00000060692
ENSRNOT00000060691 |
Nfx1
|
nuclear transcription factor, X-box binding 1 |
| chr12_+_23514981 | 0.06 |
ENSRNOT00000001937
|
Prkrip1
|
Prkr interacting protein 1 (IL11 inducible) |
| chr1_-_218532118 | 0.06 |
ENSRNOT00000086385
ENSRNOT00000018487 |
Ighmbp2
|
immunoglobulin mu binding protein 2 |
| chr10_+_70520206 | 0.06 |
ENSRNOT00000088198
ENSRNOT00000090446 ENSRNOT00000085799 |
Ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr2_-_30340103 | 0.06 |
ENSRNOT00000024023
ENSRNOT00000067722 |
Bdp1
|
B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB |
| chr6_-_1622196 | 0.06 |
ENSRNOT00000007492
|
Prkd3
|
protein kinase D3 |
| chr7_-_99677268 | 0.06 |
ENSRNOT00000013315
|
RGD1564420
|
similar to Hypothetical protein MGC31278 |
| chr9_+_98621506 | 0.06 |
ENSRNOT00000056612
ENSRNOT00000081992 |
Traf3ip1
|
TRAF3 interacting protein 1 |
| chr12_+_17277635 | 0.06 |
ENSRNOT00000001734
|
LOC498154
|
hypothetical protein LOC498154 |
| chr16_+_74237001 | 0.06 |
ENSRNOT00000026039
|
Polb
|
DNA polymerase beta |
| chr1_-_227002139 | 0.06 |
ENSRNOT00000028405
|
Ccdc86
|
coiled-coil domain containing 86 |
| chr5_+_156850206 | 0.06 |
ENSRNOT00000021833
|
Mul1
|
mitochondrial E3 ubiquitin protein ligase 1 |
| chr7_+_117773230 | 0.06 |
ENSRNOT00000077562
|
Lrrc14
|
leucine rich repeat containing 14 |
| chr5_-_57008795 | 0.06 |
ENSRNOT00000090891
ENSRNOT00000046463 |
Aptx
|
aprataxin |
| chr6_+_135610743 | 0.06 |
ENSRNOT00000010906
|
Traf3
|
Tnf receptor-associated factor 3 |
| chr8_+_36410683 | 0.06 |
ENSRNOT00000015177
|
Srpra
|
SRP receptor alpha subunit |
| chr3_+_7279340 | 0.06 |
ENSRNOT00000017029
|
Ak8
|
adenylate kinase 8 |
| chr11_-_61748768 | 0.06 |
ENSRNOT00000078879
|
Ccdc191
|
coiled-coil domain containing 191 |
| chr20_-_10257044 | 0.05 |
ENSRNOT00000068289
|
Wdr4
|
WD repeat domain 4 |
| chr5_+_134593517 | 0.05 |
ENSRNOT00000013471
|
Efcab14
|
EF-hand calcium binding domain 14 |
| chr12_+_504007 | 0.05 |
ENSRNOT00000001475
|
Brca2
|
BRCA2, DNA repair associated |
| chr20_+_38935820 | 0.05 |
ENSRNOT00000001059
|
Hsf2
|
heat shock transcription factor 2 |
| chr4_+_99399148 | 0.05 |
ENSRNOT00000088018
|
Rnf103
|
ring finger protein 103 |
| chr7_+_27081667 | 0.05 |
ENSRNOT00000066143
|
Nfyb
|
nuclear transcription factor Y subunit beta |
| chr12_-_47031498 | 0.05 |
ENSRNOT00000075320
|
Triap1
|
TP53 regulated inhibitor of apoptosis 1 |
| chrX_-_123474154 | 0.05 |
ENSRNOT00000092415
ENSRNOT00000092455 |
RGD1564541
|
similar to hypothetical protein FLJ22965 |
| chr16_-_32439421 | 0.05 |
ENSRNOT00000043100
|
Nek1
|
NIMA-related kinase 1 |
| chrX_+_158978755 | 0.05 |
ENSRNOT00000066809
|
Slc9a6
|
solute carrier family 9 member A6 |
| chr1_-_63293635 | 0.05 |
ENSRNOT00000090337
|
Zik1
|
zinc finger protein interacting with K protein 1 |
| chr10_+_4536180 | 0.05 |
ENSRNOT00000003309
|
Zc3h7a
|
zinc finger CCCH type containing 7 A |
| chr7_-_24004774 | 0.05 |
ENSRNOT00000007549
|
Pwp1
|
PWP1 homolog, endonuclein |
| chr1_-_68269117 | 0.04 |
ENSRNOT00000072367
|
LOC100911224
|
zinc finger protein interacting with ribonucleoprotein K-like |
| chr8_+_132204604 | 0.04 |
ENSRNOT00000084725
|
Exosc7
|
exosome component 7 |
| chr12_+_49328977 | 0.04 |
ENSRNOT00000000898
|
Sgsm1
|
small G protein signaling modulator 1 |
| chr4_+_138855641 | 0.04 |
ENSRNOT00000008526
|
Trnt1
|
tRNA nucleotidyl transferase 1 |
| chr9_-_10757720 | 0.04 |
ENSRNOT00000083848
|
Uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr6_+_132090218 | 0.04 |
ENSRNOT00000083208
|
Ccnk
|
cyclin K |
| chr13_+_67611708 | 0.04 |
ENSRNOT00000063833
|
Tpr
|
translocated promoter region, nuclear basket protein |
| chr17_+_6840463 | 0.04 |
ENSRNOT00000061233
|
Ubqln1
|
ubiquilin 1 |
| chr15_+_24056383 | 0.04 |
ENSRNOT00000015120
|
Socs4
|
suppressor of cytokine signaling 4 |
| chr6_+_52751106 | 0.04 |
ENSRNOT00000014373
|
Twistnb
|
TWIST neighbor |
| chr3_+_111097326 | 0.04 |
ENSRNOT00000018717
|
Vps18
|
VPS18 CORVET/HOPS core subunit |
| chrX_-_62698830 | 0.04 |
ENSRNOT00000076359
|
Pola1
|
DNA polymerase alpha 1, catalytic subunit |
| chr11_-_72929003 | 0.04 |
ENSRNOT00000071483
|
Ncbp2
|
nuclear cap binding protein subunit 2 |
| chrX_-_1786978 | 0.04 |
ENSRNOT00000011317
|
Rbm10
|
RNA binding motif protein 10 |
| chr14_+_2100106 | 0.04 |
ENSRNOT00000000064
|
Gak
|
cyclin G associated kinase |
| chr17_-_84830185 | 0.03 |
ENSRNOT00000040697
|
Skida1
|
SKI/DACH domain containing 1 |
| chr5_-_166133274 | 0.03 |
ENSRNOT00000078830
|
Kif1b
|
kinesin family member 1B |
| chr6_+_132090633 | 0.03 |
ENSRNOT00000057171
|
Ccnk
|
cyclin K |
| chr1_-_102849430 | 0.03 |
ENSRNOT00000086856
|
Saa4
|
serum amyloid A4 |
| chr9_-_44456554 | 0.03 |
ENSRNOT00000080011
|
Tsga10
|
testis specific 10 |
| chr16_+_6712389 | 0.03 |
ENSRNOT00000035542
|
Rft1
|
RFT1 homolog |
| chr5_+_117583502 | 0.03 |
ENSRNOT00000010933
|
Usp1
|
ubiquitin specific peptidase 1 |
| chr10_+_38104558 | 0.03 |
ENSRNOT00000067809
|
Fstl4
|
follistatin-like 4 |
| chr11_-_64752544 | 0.03 |
ENSRNOT00000048738
|
Tmem39a
|
transmembrane protein 39a |
| chr20_-_4530126 | 0.03 |
ENSRNOT00000000481
|
Skiv2l
|
Ski2 like RNA helicase |
| chr15_-_34431325 | 0.03 |
ENSRNOT00000027511
|
Dhrs1
|
dehydrogenase/reductase 1 |
| chr9_-_65916949 | 0.03 |
ENSRNOT00000077483
|
Tmem237
|
transmembrane protein 237 |
| chr1_-_221420115 | 0.03 |
ENSRNOT00000028475
|
Mrpl49
|
mitochondrial ribosomal protein L49 |
| chr4_-_7529106 | 0.03 |
ENSRNOT00000013865
|
Nupl2
|
nucleoporin like 2 |
| chr8_-_132032971 | 0.03 |
ENSRNOT00000005502
|
RGD1311745
|
similar to RIKEN cDNA 1110059G10 |
| chr3_+_138715570 | 0.03 |
ENSRNOT00000064723
|
Sec23b
|
Sec23 homolog B, coat complex II component |
| chr12_+_24669449 | 0.03 |
ENSRNOT00000044449
|
Wbscr22
|
Williams Beuren syndrome chromosome region 22 |
| chr8_-_128009951 | 0.03 |
ENSRNOT00000018056
|
Oxsr1
|
oxidative-stress responsive 1 |
| chr10_-_68142864 | 0.03 |
ENSRNOT00000004609
|
Myo1d
|
myosin ID |
| chr2_+_122782060 | 0.03 |
ENSRNOT00000019108
ENSRNOT00000089353 |
Acad9
|
acyl-CoA dehydrogenase family, member 9 |
| chr12_-_48663143 | 0.03 |
ENSRNOT00000082809
|
Ficd
|
FIC domain containing |
| chr6_-_99400822 | 0.03 |
ENSRNOT00000089594
|
Zbtb25
|
zinc finger and BTB domain containing 25 |
| chr7_-_135850542 | 0.02 |
ENSRNOT00000038032
|
Twf1
|
twinfilin actin-binding protein 1 |
| chr13_-_26850460 | 0.02 |
ENSRNOT00000003715
|
Vps4b
|
vacuolar protein sorting 4 homolog B |
| chr17_+_11856525 | 0.02 |
ENSRNOT00000014546
|
Sptlc1
|
serine palmitoyltransferase, long chain base subunit 1 |
| chrX_+_159513800 | 0.02 |
ENSRNOT00000065636
|
Htatsf1
|
HIV-1 Tat specific factor 1 |
| chr19_+_31867981 | 0.02 |
ENSRNOT00000024753
|
Abce1
|
ATP binding cassette subfamily E member 1 |
| chr19_+_43251539 | 0.02 |
ENSRNOT00000024793
|
Ddx19a
|
DEAD-box helicase 19A |
| chr4_-_51946715 | 0.02 |
ENSRNOT00000079130
|
Pot1
|
protection of telomeres 1 |
| chr7_-_64341207 | 0.02 |
ENSRNOT00000006051
|
Tmem5
|
transmembrane protein 5 |
| chr19_+_25262076 | 0.02 |
ENSRNOT00000030658
|
RGD1306072
|
hypothetical LOC304654 |
| chr1_-_219863926 | 0.02 |
ENSRNOT00000026454
ENSRNOT00000066455 |
Rce1
|
Ras converting CAAX endopeptidase 1 |
| chr4_+_7122890 | 0.02 |
ENSRNOT00000076339
ENSRNOT00000076418 ENSRNOT00000014718 |
Abcf2
|
ATP binding cassette subfamily F member 2 |
| chr2_+_264864351 | 0.02 |
ENSRNOT00000015434
|
Lrrc40
|
leucine rich repeat containing 40 |
| chr12_-_37444282 | 0.02 |
ENSRNOT00000001368
|
Gtf2h3
|
general transcription factor IIH subunit 3 |
| chr19_-_31867850 | 0.02 |
ENSRNOT00000024689
|
Anapc10
|
anaphase promoting complex subunit 10 |
| chr2_+_27365148 | 0.02 |
ENSRNOT00000021549
|
Col4a3bp
|
collagen type IV alpha 3 binding protein |
| chr6_-_77243677 | 0.02 |
ENSRNOT00000011419
|
Mbip
|
MAP3K12 binding inhibitory protein 1 |
| chr14_+_36216002 | 0.02 |
ENSRNOT00000038506
|
Scfd2
|
sec1 family domain containing 2 |
| chr1_+_224957517 | 0.02 |
ENSRNOT00000026033
|
Nxf1
|
nuclear RNA export factor 1 |
| chr1_+_221099998 | 0.02 |
ENSRNOT00000028262
|
Ltbp3
|
latent transforming growth factor beta binding protein 3 |
| chr5_+_148320438 | 0.02 |
ENSRNOT00000018742
|
Pef1
|
penta-EF hand domain containing 1 |
| chr10_-_38969501 | 0.02 |
ENSRNOT00000090691
ENSRNOT00000081309 ENSRNOT00000010029 |
Il4
|
interleukin 4 |
| chr3_+_58443101 | 0.02 |
ENSRNOT00000002075
|
Itga6
|
integrin subunit alpha 6 |
| chr3_-_93698121 | 0.02 |
ENSRNOT00000012094
ENSRNOT00000044143 |
Nat10
|
N-acetyltransferase 10 |
| chr12_-_24669464 | 0.02 |
ENSRNOT00000043393
|
Dnajc30
|
DnaJ heat shock protein family (Hsp40) member C30 |
| chr14_+_100415668 | 0.01 |
ENSRNOT00000008057
|
C1d
|
C1D nuclear receptor co-repressor |
| chr2_-_211638788 | 0.01 |
ENSRNOT00000027666
|
Stxbp3
|
syntaxin binding protein 3 |
| chr5_+_76150840 | 0.01 |
ENSRNOT00000020702
|
Dnajc25
|
DnaJ heat shock protein family (Hsp40) member C25 |
| chr7_-_31872423 | 0.01 |
ENSRNOT00000012715
|
Tmpo
|
thymopoietin |
Network of associatons between targets according to the STRING database.
First level regulatory network of Creb1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 0.2 | GO:1990790 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 0.1 | 0.2 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.1 | 0.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.2 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.1 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) |
| 0.0 | 0.2 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.2 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.1 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.1 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.0 | 0.2 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.4 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.1 | GO:0031632 | positive regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031632) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.2 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.0 | 0.4 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.1 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.0 | 0.2 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0090172 | meiotic metaphase I plate congression(GO:0043060) microtubule cytoskeleton organization involved in homologous chromosome segregation(GO:0090172) |
| 0.0 | 0.1 | GO:1990928 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) response to amino acid starvation(GO:1990928) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.3 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:1904925 | negative regulation of defense response to virus by host(GO:0050689) positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.2 | GO:0044827 | modulation by host of viral genome replication(GO:0044827) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 0.2 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.0 | GO:0061738 | late endosomal microautophagy(GO:0061738) |
| 0.0 | 0.0 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:0042148 | meiotic DNA recombinase assembly(GO:0000707) strand invasion(GO:0042148) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.1 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.4 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.1 | GO:0002945 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.0 | GO:0055087 | Ski complex(GO:0055087) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.3 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0016890 | site-specific endodeoxyribonuclease activity, specific for altered base(GO:0016890) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.2 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.0 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.4 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |