Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
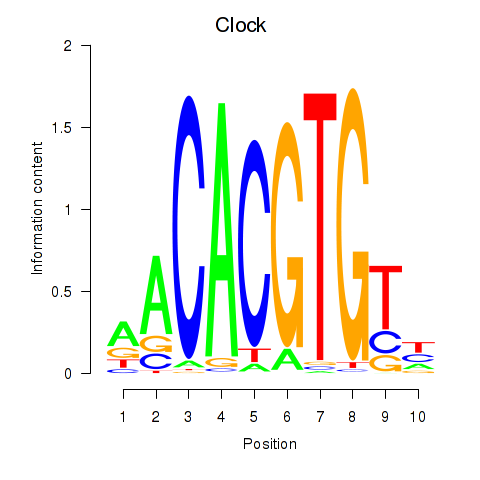
Results for Clock
Z-value: 0.28
Transcription factors associated with Clock
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Clock
|
ENSRNOG00000002175 | clock circadian regulator |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Clock | rn6_v1_chr14_+_34455934_34455934 | -0.72 | 1.7e-01 | Click! |
Activity profile of Clock motif
Sorted Z-values of Clock motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_221773254 | 0.08 |
ENSRNOT00000028646
|
Rasgrp2
|
RAS guanyl releasing protein 2 |
| chr1_-_265506046 | 0.07 |
ENSRNOT00000023963
|
Npm3
|
nucleophosmin/nucleoplasmin, 3 |
| chr8_-_130127392 | 0.07 |
ENSRNOT00000026159
|
Cck
|
cholecystokinin |
| chr5_+_165405168 | 0.07 |
ENSRNOT00000014934
|
Srm
|
spermidine synthase |
| chr8_-_62386264 | 0.06 |
ENSRNOT00000026165
|
Cplx3
|
complexin 3 |
| chr6_+_42852683 | 0.05 |
ENSRNOT00000079185
|
Odc1
|
ornithine decarboxylase 1 |
| chr16_+_49462889 | 0.05 |
ENSRNOT00000039909
|
Ankrd37
|
ankyrin repeat domain 37 |
| chr7_-_82059247 | 0.05 |
ENSRNOT00000087177
|
Rspo2
|
R-spondin 2 |
| chr10_-_88667345 | 0.05 |
ENSRNOT00000025518
|
Kcnh4
|
potassium voltage-gated channel subfamily H member 4 |
| chr2_+_127538659 | 0.05 |
ENSRNOT00000093483
ENSRNOT00000058476 |
Slc25a31
|
solute carrier family 25 member 31 |
| chr3_+_14588870 | 0.05 |
ENSRNOT00000071712
|
LOC100912604
|
spermidine synthase-like |
| chr8_-_52814188 | 0.04 |
ENSRNOT00000087265
ENSRNOT00000047633 |
Rexo2
|
RNA exonuclease 2 |
| chr5_+_74649765 | 0.04 |
ENSRNOT00000075952
|
Palm2
|
paralemmin 2 |
| chr10_-_83898527 | 0.04 |
ENSRNOT00000009815
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr5_+_152680407 | 0.04 |
ENSRNOT00000076864
|
Stmn1
|
stathmin 1 |
| chr6_-_114488880 | 0.04 |
ENSRNOT00000087560
|
AC118957.1
|
|
| chr18_+_72550219 | 0.04 |
ENSRNOT00000046847
|
Smad2
|
SMAD family member 2 |
| chr2_+_187893875 | 0.04 |
ENSRNOT00000093014
|
Mex3a
|
mex-3 RNA binding family member A |
| chr1_-_198120061 | 0.04 |
ENSRNOT00000026231
|
Slx1b
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr2_-_187160373 | 0.03 |
ENSRNOT00000018961
|
Ntrk1
|
neurotrophic receptor tyrosine kinase 1 |
| chr6_+_33786627 | 0.03 |
ENSRNOT00000008359
|
Pum2
|
pumilio RNA-binding family member 2 |
| chr20_-_43108198 | 0.03 |
ENSRNOT00000000742
|
Hdac2
|
histone deacetylase 2 |
| chr1_+_198120099 | 0.03 |
ENSRNOT00000073652
|
Bola2
|
bolA family member 2 |
| chr12_-_3924415 | 0.03 |
ENSRNOT00000067752
|
AABR07035074.1
|
|
| chr9_+_17817721 | 0.03 |
ENSRNOT00000086986
ENSRNOT00000026920 |
Hsp90ab1
|
heat shock protein 90 alpha family class B member 1 |
| chr19_+_58565576 | 0.03 |
ENSRNOT00000026965
|
Ntpcr
|
nucleoside-triphosphatase, cancer-related |
| chr6_+_43829945 | 0.03 |
ENSRNOT00000086548
|
Klf11
|
Kruppel-like factor 11 |
| chr5_+_140979435 | 0.03 |
ENSRNOT00000056592
|
Pabpc4
|
poly(A) binding protein, cytoplasmic 4 |
| chr10_+_89236256 | 0.03 |
ENSRNOT00000027957
|
Psme3
|
proteasome activator subunit 3 |
| chr3_+_71020534 | 0.02 |
ENSRNOT00000007276
|
Zc3h15
|
zinc finger CCCH-type containing 15 |
| chr11_-_83985385 | 0.02 |
ENSRNOT00000002334
|
LOC102551435
|
endothelin-converting enzyme 2-like |
| chr5_-_147953093 | 0.02 |
ENSRNOT00000075270
|
Khdrbs1
|
KH RNA binding domain containing, signal transduction associated 1 |
| chr1_-_163328591 | 0.02 |
ENSRNOT00000034843
|
Tsku
|
tsukushi, small leucine rich proteoglycan |
| chr2_-_148891900 | 0.02 |
ENSRNOT00000018488
|
Siah2
|
siah E3 ubiquitin protein ligase 2 |
| chr10_+_54246250 | 0.02 |
ENSRNOT00000004880
|
Rcvrn
|
recoverin |
| chr3_+_113415774 | 0.02 |
ENSRNOT00000056151
|
Serf2
|
small EDRK-rich factor 2 |
| chr8_-_115140080 | 0.02 |
ENSRNOT00000015851
|
Acy1
|
aminoacylase 1 |
| chr9_+_66495832 | 0.02 |
ENSRNOT00000022676
ENSRNOT00000091283 |
Nop58
|
NOP58 ribonucleoprotein |
| chr7_-_27240528 | 0.02 |
ENSRNOT00000029435
ENSRNOT00000079731 |
Hsp90b1
|
heat shock protein 90 beta family member 1 |
| chr13_+_100129761 | 0.02 |
ENSRNOT00000038340
|
Eif3el1
|
eukaryotic translation initiation factor 3, subunit E-like 1 |
| chr4_-_78879294 | 0.02 |
ENSRNOT00000084543
|
Igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr11_-_60613718 | 0.02 |
ENSRNOT00000002906
|
Atg3
|
autophagy related 3 |
| chr3_+_61685619 | 0.02 |
ENSRNOT00000002140
|
Hoxd1
|
homeo box D1 |
| chrX_+_16946207 | 0.01 |
ENSRNOT00000003981
|
RGD1563606
|
similar to cysteine-rich protein 2 |
| chr10_-_57618527 | 0.01 |
ENSRNOT00000037517
|
C1qbp
|
complement C1q binding protein |
| chr13_-_90814119 | 0.01 |
ENSRNOT00000011208
|
Tagln2
|
transgelin 2 |
| chr18_+_36829062 | 0.01 |
ENSRNOT00000025467
ENSRNOT00000086979 |
Tcerg1
|
transcription elongation regulator 1 |
| chr10_-_11760620 | 0.01 |
ENSRNOT00000009283
|
Dnase1
|
deoxyribonuclease 1 |
| chr19_+_53055745 | 0.01 |
ENSRNOT00000074430
|
Foxl1
|
forkhead box L1 |
| chr6_-_55001464 | 0.01 |
ENSRNOT00000006618
|
Ahr
|
aryl hydrocarbon receptor |
| chr10_-_40375605 | 0.01 |
ENSRNOT00000014464
|
Anxa6
|
annexin A6 |
| chr10_+_83476107 | 0.01 |
ENSRNOT00000075733
|
Phb
|
prohibitin |
| chr10_+_57057608 | 0.01 |
ENSRNOT00000085810
ENSRNOT00000026335 |
Med11
|
mediator complex subunit 11 |
| chr7_+_77966722 | 0.01 |
ENSRNOT00000006142
|
Cthrc1
|
collagen triple helix repeat containing 1 |
| chr10_+_82352390 | 0.01 |
ENSRNOT00000004941
|
Lrrc59
|
leucine rich repeat containing 59 |
| chr4_-_77489535 | 0.01 |
ENSRNOT00000008728
|
Pdia4
|
protein disulfide isomerase family A, member 4 |
| chr8_+_44047592 | 0.01 |
ENSRNOT00000085084
|
Zfp202
|
zinc finger protein 202 |
| chr2_-_197991198 | 0.01 |
ENSRNOT00000056322
|
Ciart
|
circadian associated repressor of transcription |
| chr3_+_122803772 | 0.01 |
ENSRNOT00000009564
|
Nop56
|
NOP56 ribonucleoprotein |
| chr11_+_47090245 | 0.01 |
ENSRNOT00000060645
|
AABR07033987.1
|
|
| chr2_-_197991574 | 0.01 |
ENSRNOT00000085632
|
Ciart
|
circadian associated repressor of transcription |
| chr3_-_159802952 | 0.01 |
ENSRNOT00000011610
|
Oser1
|
oxidative stress responsive serine-rich 1 |
| chr10_-_32471454 | 0.01 |
ENSRNOT00000003224
|
Sgcd
|
sarcoglycan, delta |
| chr1_+_127802978 | 0.01 |
ENSRNOT00000055877
|
Adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr14_+_36216002 | 0.01 |
ENSRNOT00000038506
|
Scfd2
|
sec1 family domain containing 2 |
| chr9_-_16979044 | 0.01 |
ENSRNOT00000073191
ENSRNOT00000025335 |
Zfp318
|
zinc finger protein 318 |
| chr12_+_21981755 | 0.01 |
ENSRNOT00000001871
|
LOC100910540
|
7SK snRNA methylphosphate capping enzyme-like |
| chr9_-_61690956 | 0.01 |
ENSRNOT00000066589
|
Hspd1
|
heat shock protein family D member 1 |
| chr6_+_80220547 | 0.01 |
ENSRNOT00000086221
ENSRNOT00000077435 ENSRNOT00000059318 ENSRNOT00000089010 |
Mia2
|
melanoma inhibitory activity 2 |
| chr10_+_91710495 | 0.01 |
ENSRNOT00000033276
|
Rprml
|
reprimo-like |
| chr14_-_33580566 | 0.01 |
ENSRNOT00000063942
|
Paics
|
phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthase |
| chr3_-_122947075 | 0.01 |
ENSRNOT00000082369
|
Pced1a
|
PC-esterase domain containing 1A |
| chr10_+_46783979 | 0.01 |
ENSRNOT00000005237
|
Gid4
|
GID complex subunit 4 |
| chr16_+_7103998 | 0.00 |
ENSRNOT00000016581
|
Pbrm1
|
polybromo 1 |
| chr5_-_58124681 | 0.00 |
ENSRNOT00000019795
|
Sigmar1
|
sigma non-opioid intracellular receptor 1 |
| chr3_-_163935617 | 0.00 |
ENSRNOT00000074023
|
Kcnb1
|
potassium voltage-gated channel subfamily B member 1 |
| chr5_+_60250546 | 0.00 |
ENSRNOT00000017707
|
Zcchc7
|
zinc finger CCHC-type containing 7 |
| chr8_+_22050222 | 0.00 |
ENSRNOT00000028096
|
Icam5
|
intercellular adhesion molecule 5 |
| chr10_-_57005272 | 0.00 |
ENSRNOT00000026102
|
Pelp1
|
proline, glutamate and leucine rich protein 1 |
| chr10_-_37455022 | 0.00 |
ENSRNOT00000016742
|
LOC103694902
|
ubiquitin-conjugating enzyme E2 B |
| chr14_-_100372760 | 0.00 |
ENSRNOT00000007824
|
Pno1
|
partner of NOB1 homolog |
| chr3_+_11424099 | 0.00 |
ENSRNOT00000019184
|
Ptges2
|
prostaglandin E synthase 2 |
| chr9_+_15393330 | 0.00 |
ENSRNOT00000075630
|
Bysl
|
bystin-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Clock
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.0 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.0 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process(GO:0009446) putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.0 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.0 | 0.0 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |