Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
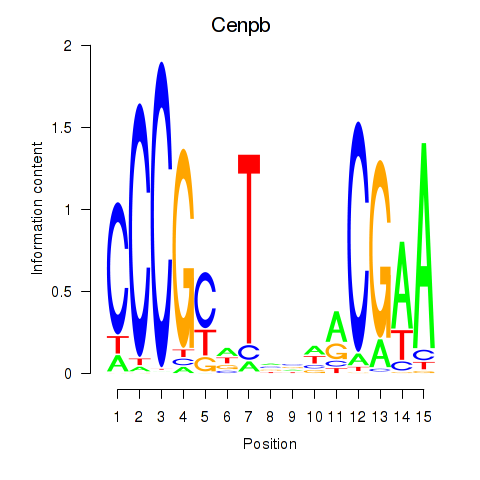
Results for Cenpb
Z-value: 0.64
Transcription factors associated with Cenpb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cenpb
|
ENSRNOG00000057284 | centromere protein B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cenpb | rn6_v1_chr3_-_123723524_123723524 | -0.40 | 5.1e-01 | Click! |
Activity profile of Cenpb motif
Sorted Z-values of Cenpb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_61332351 | 0.21 |
ENSRNOT00000014492
|
Nrp1
|
neuropilin 1 |
| chr1_-_103014388 | 0.21 |
ENSRNOT00000045719
|
Uevld
|
UEV and lactate/malate dehyrogenase domains |
| chr8_-_116531784 | 0.20 |
ENSRNOT00000024529
|
Rbm5
|
RNA binding motif protein 5 |
| chr3_-_146470293 | 0.20 |
ENSRNOT00000009627
|
Acss1
|
acyl-CoA synthetase short-chain family member 1 |
| chr6_+_86785771 | 0.19 |
ENSRNOT00000066702
|
Prpf39
|
pre-mRNA processing factor 39 |
| chr3_+_64190973 | 0.19 |
ENSRNOT00000011342
|
AABR07052587.1
|
|
| chr3_+_176821652 | 0.18 |
ENSRNOT00000055030
|
Rtel1
|
regulator of telomere elongation helicase 1 |
| chr8_-_116532169 | 0.18 |
ENSRNOT00000085364
|
Rbm5
|
RNA binding motif protein 5 |
| chr18_+_27558089 | 0.17 |
ENSRNOT00000027499
|
Fam53c
|
family with sequence similarity 53, member C |
| chrX_+_74315850 | 0.17 |
ENSRNOT00000076538
|
LOC680227
|
LRRGT00193 |
| chr6_-_65319527 | 0.16 |
ENSRNOT00000005618
|
Stxbp6
|
syntaxin binding protein 6 |
| chrX_+_156873849 | 0.16 |
ENSRNOT00000085410
|
Arhgap4
|
Rho GTPase activating protein 4 |
| chr9_-_103207190 | 0.16 |
ENSRNOT00000026010
|
St8sia4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr5_+_147476221 | 0.16 |
ENSRNOT00000010730
|
Sync
|
syncoilin, intermediate filament protein |
| chr1_+_267618565 | 0.16 |
ENSRNOT00000076251
|
Gsto2
|
glutathione S-transferase omega 2 |
| chrX_+_104882704 | 0.15 |
ENSRNOT00000079572
ENSRNOT00000074330 ENSRNOT00000082983 |
Cstf2
|
cleavage stimulation factor subunit 2 |
| chr8_-_84632817 | 0.15 |
ENSRNOT00000076942
|
Mlip
|
muscular LMNA-interacting protein |
| chr16_-_3139177 | 0.15 |
ENSRNOT00000039311
|
Ccdc66
|
coiled-coil domain containing 66 |
| chr15_+_51756978 | 0.15 |
ENSRNOT00000024067
|
Egr3
|
early growth response 3 |
| chr15_+_83442144 | 0.14 |
ENSRNOT00000039344
|
Bora
|
bora, aurora kinase A activator |
| chr9_-_104350308 | 0.14 |
ENSRNOT00000033958
|
Slco4c1
|
solute carrier organic anion transporter family, member 4C1 |
| chr9_+_47281961 | 0.14 |
ENSRNOT00000065234
|
Slc9a4
|
solute carrier family 9 member A4 |
| chr9_-_50884596 | 0.14 |
ENSRNOT00000016285
|
Kdelc1
|
KDEL motif containing 1 |
| chr18_-_73873280 | 0.13 |
ENSRNOT00000075580
|
Rnf165
|
ring finger protein 165 |
| chr14_-_112946875 | 0.13 |
ENSRNOT00000081981
|
LOC103690141
|
coiled-coil domain-containing protein 85A-like |
| chr2_+_205553163 | 0.13 |
ENSRNOT00000039572
|
Nras
|
neuroblastoma RAS viral oncogene homolog |
| chr10_-_47453442 | 0.13 |
ENSRNOT00000050061
|
Usp22
|
ubiquitin specific peptidase 22 |
| chr3_-_3824284 | 0.13 |
ENSRNOT00000068110
ENSRNOT00000061682 |
Snapc4
|
small nuclear RNA activating complex, polypeptide 4 |
| chr16_-_14348046 | 0.13 |
ENSRNOT00000018630
|
Cdhr1
|
cadherin-related family member 1 |
| chr9_-_92530938 | 0.12 |
ENSRNOT00000064875
|
Slc16a14
|
solute carrier family 16, member 14 |
| chr2_+_211176556 | 0.12 |
ENSRNOT00000055880
|
Psrc1
|
proline and serine rich coiled-coil 1 |
| chr18_+_60639860 | 0.12 |
ENSRNOT00000083363
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr11_+_84396033 | 0.12 |
ENSRNOT00000002316
|
Abcc5
|
ATP binding cassette subfamily C member 5 |
| chr3_+_8958090 | 0.12 |
ENSRNOT00000086789
ENSRNOT00000064557 |
Dolpp1
|
dolichyldiphosphatase 1 |
| chr10_+_29606748 | 0.11 |
ENSRNOT00000080720
|
AABR07029467.2
|
|
| chr20_-_47306318 | 0.11 |
ENSRNOT00000075151
|
Nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr2_-_33841499 | 0.11 |
ENSRNOT00000040533
|
Srek1
|
splicing regulatory glutamic acid and lysine rich protein 1 |
| chr7_-_98098268 | 0.11 |
ENSRNOT00000010361
|
Fbxo32
|
F-box protein 32 |
| chrX_+_39711201 | 0.11 |
ENSRNOT00000080512
ENSRNOT00000009802 |
Cnksr2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr16_-_32868680 | 0.11 |
ENSRNOT00000015974
ENSRNOT00000082392 |
Aadat
|
aminoadipate aminotransferase |
| chr18_-_70924708 | 0.11 |
ENSRNOT00000025257
|
Lipg
|
lipase G, endothelial type |
| chr10_+_36741434 | 0.11 |
ENSRNOT00000064078
|
Col23a1
|
collagen type XXIII alpha 1 chain |
| chr20_-_6257604 | 0.11 |
ENSRNOT00000092489
|
Stk38
|
serine/threonine kinase 38 |
| chr1_-_128695995 | 0.11 |
ENSRNOT00000077020
|
Synm
|
synemin |
| chr1_-_31881531 | 0.10 |
ENSRNOT00000081225
|
Tppp
|
tubulin polymerization promoting protein |
| chr7_-_118396728 | 0.10 |
ENSRNOT00000066431
|
Rbfox2
|
RNA binding protein, fox-1 homolog 2 |
| chr1_-_165967069 | 0.10 |
ENSRNOT00000089359
|
Arhgef17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr3_-_170955399 | 0.10 |
ENSRNOT00000084990
|
Bmp7
|
bone morphogenetic protein 7 |
| chr16_+_20039969 | 0.10 |
ENSRNOT00000024866
|
Colgalt1
|
collagen beta(1-O)galactosyltransferase 1 |
| chr10_+_89578212 | 0.10 |
ENSRNOT00000028178
|
Arl4d
|
ADP-ribosylation factor like GTPase 4D |
| chr10_+_94988362 | 0.10 |
ENSRNOT00000066525
|
Cep95
|
centrosomal protein 95 |
| chr10_-_105368242 | 0.10 |
ENSRNOT00000075293
ENSRNOT00000072230 |
Rnf157
|
ring finger protein 157 |
| chr5_-_160070916 | 0.09 |
ENSRNOT00000067517
ENSRNOT00000055791 |
Spen
|
spen family transcriptional repressor |
| chr12_+_12374790 | 0.09 |
ENSRNOT00000001347
|
Tecpr1
|
tectonin beta-propeller repeat containing 1 |
| chr1_-_18058055 | 0.09 |
ENSRNOT00000020988
|
Ptprk
|
protein tyrosine phosphatase, receptor type, K |
| chr2_-_219628997 | 0.09 |
ENSRNOT00000064484
|
Trmt13
|
tRNA methyltransferase 13 homolog |
| chr4_-_150506406 | 0.09 |
ENSRNOT00000076307
|
Zfp248
|
zinc finger protein 248 |
| chr3_+_149935731 | 0.09 |
ENSRNOT00000021907
|
Cbfa2t2
|
CBFA2/RUNX1 translocation partner 2 |
| chr10_+_89181180 | 0.08 |
ENSRNOT00000078931
ENSRNOT00000027770 |
Wnk4
|
WNK lysine deficient protein kinase 4 |
| chr1_-_88112683 | 0.08 |
ENSRNOT00000090615
|
Spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr6_+_43234526 | 0.08 |
ENSRNOT00000086808
|
Asap2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
| chr7_-_11257977 | 0.08 |
ENSRNOT00000027932
|
Tbxa2r
|
thromboxane A2 receptor |
| chr7_+_14891001 | 0.08 |
ENSRNOT00000041034
|
Cyp4f4
|
cytochrome P450, family 4, subfamily f, polypeptide 4 |
| chr10_-_90151042 | 0.08 |
ENSRNOT00000055187
|
Hdac5
|
histone deacetylase 5 |
| chr10_-_49009156 | 0.08 |
ENSRNOT00000072970
|
Mapk7
|
mitogen-activated protein kinase 7 |
| chr20_-_29184185 | 0.08 |
ENSRNOT00000090771
|
Mcu
|
mitochondrial calcium uniporter |
| chr1_+_72956026 | 0.08 |
ENSRNOT00000031462
|
Rdh13
|
retinol dehydrogenase 13 |
| chr10_+_59613398 | 0.08 |
ENSRNOT00000025553
|
Ncbp3
|
nuclear cap binding subunit 3 |
| chr9_+_71230108 | 0.08 |
ENSRNOT00000018326
|
Creb1
|
cAMP responsive element binding protein 1 |
| chrX_+_76786466 | 0.08 |
ENSRNOT00000090665
|
Fgf16
|
fibroblast growth factor 16 |
| chrX_-_105622156 | 0.08 |
ENSRNOT00000029511
|
Armcx2
|
armadillo repeat containing, X-linked 2 |
| chr20_+_3156170 | 0.08 |
ENSRNOT00000082880
|
RT1-S2
|
RT1 class Ib, locus S2 |
| chr1_-_219519398 | 0.08 |
ENSRNOT00000025546
|
Ssh3
|
slingshot protein phosphatase 3 |
| chr4_-_147756294 | 0.08 |
ENSRNOT00000014537
|
Mbd4
|
methyl-CpG binding domain 4 DNA glycosylase |
| chr2_+_27949208 | 0.08 |
ENSRNOT00000052072
|
Gfm2
|
G elongation factor, mitochondrial 2 |
| chrX_-_1784807 | 0.08 |
ENSRNOT00000077453
|
Rbm10
|
RNA binding motif protein 10 |
| chr9_+_66568074 | 0.08 |
ENSRNOT00000035238
|
Bmpr2
|
bone morphogenetic protein receptor type 2 |
| chr7_-_11824742 | 0.07 |
ENSRNOT00000051467
|
Dot1l
|
DOT1 like histone lysine methyltransferase |
| chr3_+_101156212 | 0.07 |
ENSRNOT00000065333
|
Ccdc34
|
coiled-coil domain containing 34 |
| chr2_+_209766512 | 0.07 |
ENSRNOT00000092240
|
Kcna3
|
potassium voltage-gated channel subfamily A member 3 |
| chr18_+_15883182 | 0.07 |
ENSRNOT00000088353
ENSRNOT00000076137 |
Zfp35
|
zinc finger protein 35 |
| chr10_+_67559385 | 0.07 |
ENSRNOT00000082685
ENSRNOT00000006218 ENSRNOT00000079696 |
Rhot1
|
ras homolog family member T1 |
| chr6_+_48866601 | 0.07 |
ENSRNOT00000077321
ENSRNOT00000079891 |
Pxdn
|
peroxidasin |
| chr14_-_104612597 | 0.07 |
ENSRNOT00000007002
|
Slc1a4
|
solute carrier family 1 member 4 |
| chr1_+_85386470 | 0.07 |
ENSRNOT00000093332
ENSRNOT00000044326 |
Plekhg2
|
pleckstrin homology and RhoGEF domain containing G2 |
| chr15_-_41338284 | 0.07 |
ENSRNOT00000077225
|
Tnfrsf19
|
TNF receptor superfamily member 19 |
| chr3_+_130008885 | 0.07 |
ENSRNOT00000070972
|
Slx4ip
|
SLX4 interacting protein |
| chr3_+_8701855 | 0.07 |
ENSRNOT00000021431
|
Tbc1d13
|
TBC1 domain family, member 13 |
| chr7_-_130128589 | 0.07 |
ENSRNOT00000079777
ENSRNOT00000009325 |
Mapk11
|
mitogen-activated protein kinase 11 |
| chr2_+_116028781 | 0.07 |
ENSRNOT00000089477
|
Phc3
|
polyhomeotic homolog 3 |
| chr16_-_48863204 | 0.07 |
ENSRNOT00000014095
|
Casp3
|
caspase 3 |
| chr3_-_6054483 | 0.07 |
ENSRNOT00000084491
|
Brd3
|
bromodomain containing 3 |
| chr1_+_261753364 | 0.07 |
ENSRNOT00000076659
|
R3hcc1l
|
R3H domain and coiled-coil containing 1-like |
| chrX_-_56765893 | 0.07 |
ENSRNOT00000076283
|
Il1rapl1
|
interleukin 1 receptor accessory protein-like 1 |
| chrX_+_75792931 | 0.07 |
ENSRNOT00000037575
|
Magee2
|
MAGE family member E2 |
| chr12_-_41485122 | 0.07 |
ENSRNOT00000001859
|
Ddx54
|
DEAD-box helicase 54 |
| chr2_-_211322719 | 0.07 |
ENSRNOT00000027493
|
RGD1310209
|
similar to KIAA1324 protein |
| chr7_+_2827247 | 0.07 |
ENSRNOT00000090689
|
Rnf41
|
ring finger protein 41 |
| chr2_+_219628695 | 0.07 |
ENSRNOT00000067324
|
Sass6
|
SAS-6 centriolar assembly protein |
| chr4_+_70903253 | 0.06 |
ENSRNOT00000019720
|
Ephb6
|
Eph receptor B6 |
| chr1_-_167308827 | 0.06 |
ENSRNOT00000027590
ENSRNOT00000027575 |
Nup98
|
nucleoporin 98 |
| chr10_-_66602987 | 0.06 |
ENSRNOT00000017949
|
Wsb1
|
WD repeat and SOCS box-containing 1 |
| chr2_+_192538899 | 0.06 |
ENSRNOT00000045691
ENSRNOT00000085931 |
LOC102550416
|
small proline-rich protein 2I-like |
| chr10_+_94260197 | 0.06 |
ENSRNOT00000063973
|
Taco1
|
translational activator of cytochrome c oxidase I |
| chr3_-_122947075 | 0.06 |
ENSRNOT00000082369
|
Pced1a
|
PC-esterase domain containing 1A |
| chr8_+_103774358 | 0.06 |
ENSRNOT00000014481
|
Xrn1
|
5'-3' exoribonuclease 1 |
| chr4_+_81311490 | 0.06 |
ENSRNOT00000016265
|
Snx10
|
sorting nexin 10 |
| chr14_-_67170361 | 0.06 |
ENSRNOT00000005477
|
Slit2
|
slit guidance ligand 2 |
| chr7_+_40217269 | 0.06 |
ENSRNOT00000082090
|
Cep290
|
centrosomal protein 290 |
| chr2_+_188844073 | 0.06 |
ENSRNOT00000028117
|
Kcnn3
|
potassium calcium-activated channel subfamily N member 3 |
| chr11_+_80742467 | 0.06 |
ENSRNOT00000002507
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr1_+_256101903 | 0.06 |
ENSRNOT00000022384
|
Hhex
|
hematopoietically expressed homeobox |
| chr17_-_78735324 | 0.06 |
ENSRNOT00000036299
|
Cdnf
|
cerebral dopamine neurotrophic factor |
| chr13_+_89586283 | 0.06 |
ENSRNOT00000079355
ENSRNOT00000049873 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr16_-_7588841 | 0.06 |
ENSRNOT00000084645
|
Mettl6
|
methyltransferase like 6 |
| chr5_-_59543101 | 0.06 |
ENSRNOT00000019532
|
Gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr15_-_28721127 | 0.06 |
ENSRNOT00000017720
|
Mettl3
|
methyltransferase-like 3 |
| chr7_+_125034764 | 0.06 |
ENSRNOT00000015767
|
Pnpla3
|
patatin-like phospholipase domain containing 3 |
| chr7_+_40217991 | 0.06 |
ENSRNOT00000085684
|
Cep290
|
centrosomal protein 290 |
| chr20_-_3401273 | 0.06 |
ENSRNOT00000089257
ENSRNOT00000078451 ENSRNOT00000001085 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr1_+_72874404 | 0.06 |
ENSRNOT00000058900
|
Dnaaf3
|
dynein, axonemal, assembly factor 3 |
| chr13_+_99136871 | 0.05 |
ENSRNOT00000078263
ENSRNOT00000004350 |
Sde2
|
SDE2 telomere maintenance homolog |
| chr18_+_30880020 | 0.05 |
ENSRNOT00000060468
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5 |
| chr9_+_10471742 | 0.05 |
ENSRNOT00000072276
|
Safb2
|
scaffold attachment factor B2 |
| chr2_-_30664163 | 0.05 |
ENSRNOT00000024801
|
Rad17
|
RAD17 checkpoint clamp loader component |
| chr6_-_50923510 | 0.05 |
ENSRNOT00000010631
|
Bcap29
|
B-cell receptor-associated protein 29 |
| chr11_+_87522971 | 0.05 |
ENSRNOT00000043545
|
Smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr4_+_5841998 | 0.05 |
ENSRNOT00000010025
|
Xrcc2
|
X-ray repair cross complementing 2 |
| chr1_+_29191192 | 0.05 |
ENSRNOT00000018718
|
Hey2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr1_-_198120061 | 0.05 |
ENSRNOT00000026231
|
Slx1b
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr13_-_37536066 | 0.05 |
ENSRNOT00000037203
|
Ddx18
|
DEAD-box helicase 18 |
| chr2_+_257425679 | 0.05 |
ENSRNOT00000066199
|
Fubp1
|
far upstream element binding protein 1 |
| chr4_-_150506557 | 0.05 |
ENSRNOT00000076927
|
Zfp248
|
zinc finger protein 248 |
| chr1_-_222495382 | 0.05 |
ENSRNOT00000028759
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr2_-_26011429 | 0.05 |
ENSRNOT00000065143
|
Arhgef28
|
Rho guanine nucleotide exchange factor 28 |
| chr1_+_219000844 | 0.05 |
ENSRNOT00000022486
|
Kmt5b
|
lysine methyltransferase 5B |
| chr1_+_214534284 | 0.05 |
ENSRNOT00000064254
|
Ap2a2
|
adaptor-related protein complex 2, alpha 2 subunit |
| chr1_+_261802223 | 0.05 |
ENSRNOT00000081765
|
R3hcc1l
|
R3H domain and coiled-coil containing 1-like |
| chr11_-_86276430 | 0.05 |
ENSRNOT00000075164
|
Hira
|
histone cell cycle regulator |
| chr4_+_147756553 | 0.05 |
ENSRNOT00000086549
ENSRNOT00000014733 |
Ift122
|
intraflagellar transport 122 |
| chr12_+_51948650 | 0.05 |
ENSRNOT00000056735
|
Ep400
|
E1A binding protein p400 |
| chr13_+_67611708 | 0.05 |
ENSRNOT00000063833
|
Tpr
|
translocated promoter region, nuclear basket protein |
| chr8_-_113675128 | 0.05 |
ENSRNOT00000017318
|
Nudt16
|
nudix hydrolase 16 |
| chr19_+_17115412 | 0.05 |
ENSRNOT00000015876
|
Rpgrip1l
|
Rpgrip1-like |
| chr17_-_36177457 | 0.05 |
ENSRNOT00000036978
|
Mboat1
|
membrane bound O-acyltransferase domain containing 1 |
| chr11_+_64761146 | 0.05 |
ENSRNOT00000051428
|
Poglut1
|
protein O-glucosyltransferase 1 |
| chr2_-_125323454 | 0.05 |
ENSRNOT00000045837
|
Ankrd50
|
ankyrin repeat domain 50 |
| chr17_+_19249952 | 0.05 |
ENSRNOT00000023140
|
Atxn1
|
ataxin 1 |
| chr1_-_101809544 | 0.05 |
ENSRNOT00000028591
|
Kcnj14
|
potassium voltage-gated channel subfamily J member 14 |
| chr19_+_37568113 | 0.05 |
ENSRNOT00000023710
|
Fam65a
|
family with sequence similarity 65, member A |
| chr5_+_147069616 | 0.04 |
ENSRNOT00000072908
|
Trim62
|
tripartite motif-containing 62 |
| chr7_-_126691232 | 0.04 |
ENSRNOT00000055909
|
Cdpf1
|
cysteine rich, DPF motif domain containing 1 |
| chr15_-_83442008 | 0.04 |
ENSRNOT00000039400
|
Mzt1
|
mitotic spindle organizing protein 1 |
| chr10_+_56824505 | 0.04 |
ENSRNOT00000067128
|
Slc16a11
|
solute carrier family 16, member 11 |
| chr2_+_121165137 | 0.04 |
ENSRNOT00000016236
|
Sox2
|
SRY box 2 |
| chr2_+_196304492 | 0.04 |
ENSRNOT00000028640
|
Lysmd1
|
LysM domain containing 1 |
| chr10_-_70515936 | 0.04 |
ENSRNOT00000082684
|
Pex12
|
peroxisomal biogenesis factor 12 |
| chr10_-_46783889 | 0.04 |
ENSRNOT00000004948
|
Atpaf2
|
ATP synthase mitochondrial F1 complex assembly factor 2 |
| chr6_+_25076147 | 0.04 |
ENSRNOT00000010419
|
Ehd3
|
EH-domain containing 3 |
| chr7_+_97984862 | 0.04 |
ENSRNOT00000008508
|
Atad2
|
ATPase family, AAA domain containing 2 |
| chr13_-_102942863 | 0.04 |
ENSRNOT00000003198
|
Mark1
|
microtubule affinity regulating kinase 1 |
| chr1_-_141451075 | 0.04 |
ENSRNOT00000033491
|
Kif7
|
kinesin family member 7 |
| chr4_-_18396035 | 0.04 |
ENSRNOT00000034692
|
Sema3a
|
semaphorin 3A |
| chr1_+_115975324 | 0.04 |
ENSRNOT00000080907
|
Atp10a
|
ATPase phospholipid transporting 10A (putative) |
| chr1_-_80544825 | 0.04 |
ENSRNOT00000057802
ENSRNOT00000040060 ENSRNOT00000067049 ENSRNOT00000052387 ENSRNOT00000073352 |
Relb
|
RELB proto-oncogene, NF-kB subunit |
| chr10_-_91500661 | 0.04 |
ENSRNOT00000037147
|
Plekhm1
|
pleckstrin homology and RUN domain containing M1 |
| chr1_-_112436794 | 0.04 |
ENSRNOT00000019971
|
Gabrg3
|
gamma-aminobutyric acid type A receptor gamma 3 subunit |
| chr3_-_9448167 | 0.04 |
ENSRNOT00000012617
|
Abl1
|
ABL proto-oncogene 1, non-receptor tyrosine kinase |
| chr4_-_178168690 | 0.04 |
ENSRNOT00000020729
|
Sox5
|
SRY box 5 |
| chr8_-_122987191 | 0.04 |
ENSRNOT00000033976
|
Gpd1l
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr20_+_50394650 | 0.04 |
ENSRNOT00000076010
ENSRNOT00000077065 ENSRNOT00000073259 |
Popdc3
|
popeye domain containing 3 |
| chr7_-_142132173 | 0.04 |
ENSRNOT00000026613
|
Csrnp2
|
cysteine and serine rich nuclear protein 2 |
| chrX_+_75382598 | 0.04 |
ENSRNOT00000033494
|
Uprt
|
uracil phosphoribosyltransferase homolog |
| chr1_+_85324079 | 0.04 |
ENSRNOT00000093693
|
Samd4b
|
sterile alpha motif domain containing 4B |
| chr17_+_55008265 | 0.04 |
ENSRNOT00000033827
|
Zfp438
|
zinc finger protein 438 |
| chrX_+_73390903 | 0.04 |
ENSRNOT00000077002
|
Zfp449
|
zinc finger protein 449 |
| chr1_+_56982821 | 0.04 |
ENSRNOT00000059845
|
Ermard
|
ER membrane-associated RNA degradation |
| chr17_-_42241066 | 0.04 |
ENSRNOT00000059570
|
Tdp2
|
tyrosyl-DNA phosphodiesterase 2 |
| chr4_+_87026530 | 0.04 |
ENSRNOT00000018425
|
Avl9
|
AVL9 cell migration associated |
| chr3_+_119698652 | 0.04 |
ENSRNOT00000088834
ENSRNOT00000066114 |
Stard7
|
StAR-related lipid transfer domain containing 7 |
| chr9_+_82741920 | 0.04 |
ENSRNOT00000027337
|
Slc4a3
|
solute carrier family 4 member 3 |
| chr18_-_37776453 | 0.04 |
ENSRNOT00000087876
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr8_-_90975599 | 0.04 |
ENSRNOT00000090139
|
Lca5
|
LCA5, lebercilin |
| chr8_+_61805677 | 0.04 |
ENSRNOT00000089575
ENSRNOT00000048318 |
Man2c1
|
mannosidase, alpha, class 2C, member 1 |
| chr2_+_227455722 | 0.03 |
ENSRNOT00000064809
|
Sec24d
|
SEC24 homolog D, COPII coat complex component |
| chr7_-_63687978 | 0.03 |
ENSRNOT00000009260
|
Tbk1
|
TANK-binding kinase 1 |
| chr20_+_40778927 | 0.03 |
ENSRNOT00000001081
|
Smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr18_+_1971506 | 0.03 |
ENSRNOT00000092027
|
Mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr3_-_60460724 | 0.03 |
ENSRNOT00000024706
|
Chrna1
|
cholinergic receptor nicotinic alpha 1 subunit |
| chr16_-_73410777 | 0.03 |
ENSRNOT00000024128
|
Sfrp1
|
secreted frizzled-related protein 1 |
| chr20_+_6973398 | 0.03 |
ENSRNOT00000041665
|
Fgd2
|
FYVE, RhoGEF and PH domain containing 2 |
| chr8_+_48699769 | 0.03 |
ENSRNOT00000044613
|
Hyou1
|
hypoxia up-regulated 1 |
| chr6_+_80108655 | 0.03 |
ENSRNOT00000006137
|
Gemin2
|
gem (nuclear organelle) associated protein 2 |
| chr4_+_89079014 | 0.03 |
ENSRNOT00000087451
|
Herc3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr1_+_80920747 | 0.03 |
ENSRNOT00000026077
|
Zfp180
|
zinc finger protein 180 |
| chr17_-_4454701 | 0.03 |
ENSRNOT00000080750
ENSRNOT00000066950 |
Dapk1
|
death associated protein kinase 1 |
| chr2_+_178679041 | 0.03 |
ENSRNOT00000013524
|
Fam198b
|
family with sequence similarity 198, member B |
| chr16_-_7250924 | 0.03 |
ENSRNOT00000025394
|
Stab1
|
stabilin 1 |
| chr8_-_61823102 | 0.03 |
ENSRNOT00000058645
|
Neil1
|
nei-like DNA glycosylase 1 |
| chr6_+_43829945 | 0.03 |
ENSRNOT00000086548
|
Klf11
|
Kruppel-like factor 11 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Cenpb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:0003250 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) |
| 0.1 | 0.2 | GO:0038190 | vestibulocochlear nerve structural organization(GO:0021649) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) renal artery morphogenesis(GO:0061441) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 0.1 | 0.2 | GO:0019541 | acetate metabolic process(GO:0006083) propionate metabolic process(GO:0019541) |
| 0.0 | 0.1 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 | 0.2 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.0 | GO:0021828 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) facioacoustic ganglion development(GO:1903375) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.1 | GO:0072134 | nephrogenic mesenchyme morphogenesis(GO:0072134) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.1 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.1 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.0 | 0.1 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.0 | 0.2 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0090024 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 0.0 | 0.1 | GO:0061010 | gall bladder development(GO:0061010) hepatoblast differentiation(GO:0061017) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0046380 | N-acetylglucosamine biosynthetic process(GO:0006045) N-acetylneuraminate biosynthetic process(GO:0046380) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.1 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.2 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.0 | GO:0010847 | MAPK import into nucleus(GO:0000189) regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.1 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.0 | 0.1 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.0 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.0 | 0.1 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.0 | GO:0002332 | transitional stage B cell differentiation(GO:0002332) transitional one stage B cell differentiation(GO:0002333) glomerular visceral epithelial cell apoptotic process(GO:1903210) positive regulation of actin filament binding(GO:1904531) positive regulation of actin binding(GO:1904618) |
| 0.0 | 0.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.0 | 0.0 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.0 | 0.1 | GO:1904429 | regulation of t-circle formation(GO:1904429) |
| 0.0 | 0.0 | GO:0035720 | intraciliary anterograde transport(GO:0035720) ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.3 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.0 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.0 | GO:0071504 | cellular response to heparin(GO:0071504) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 0.0 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.0 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.1 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.0 | 0.0 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.0 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.2 | GO:0030613 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.1 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0015292 | uniporter activity(GO:0015292) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.1 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0009384 | N-acylmannosamine kinase activity(GO:0009384) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.0 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.0 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |