Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
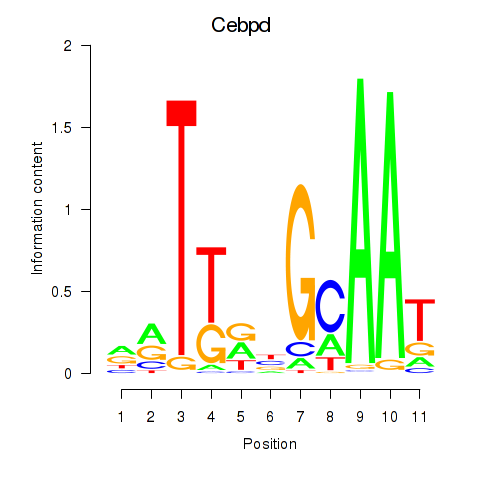
Results for Cebpd
Z-value: 0.17
Transcription factors associated with Cebpd
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cebpd
|
ENSRNOG00000050869 | CCAAT/enhancer binding protein delta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cebpd | rn6_v1_chr11_+_89008008_89008008 | 0.37 | 5.4e-01 | Click! |
Activity profile of Cebpd motif
Sorted Z-values of Cebpd motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_70871066 | 0.08 |
ENSRNOT00000015139
|
Ccl3
|
C-C motif chemokine ligand 3 |
| chr1_+_168964202 | 0.08 |
ENSRNOT00000089102
|
LOC103694855
|
hemoglobin subunit beta-2-like |
| chr9_-_78969013 | 0.06 |
ENSRNOT00000019772
ENSRNOT00000057585 |
Fn1
|
fibronectin 1 |
| chr8_-_111721303 | 0.05 |
ENSRNOT00000045628
ENSRNOT00000012725 |
Tf
|
transferrin |
| chr10_-_59112788 | 0.03 |
ENSRNOT00000041886
|
Spns3
|
SPNS sphingolipid transporter 3 |
| chr1_+_89215266 | 0.02 |
ENSRNOT00000093612
ENSRNOT00000084799 |
Dmkn
|
dermokine |
| chrX_+_33443186 | 0.02 |
ENSRNOT00000005622
|
S100g
|
S100 calcium binding protein G |
| chrX_+_143097525 | 0.01 |
ENSRNOT00000004559
|
F9
|
coagulation factor IX |
| chr3_-_160573978 | 0.01 |
ENSRNOT00000018386
|
Wfdc5
|
WAP four-disulfide core domain 5 |
| chr6_-_107678156 | 0.01 |
ENSRNOT00000014158
|
Elmsan1
|
ELM2 and Myb/SANT domain containing 1 |
| chr6_-_125723732 | 0.01 |
ENSRNOT00000084815
|
Fbln5
|
fibulin 5 |
| chr3_+_170399302 | 0.01 |
ENSRNOT00000035245
|
Cass4
|
Cas scaffolding protein family member 4 |
| chr16_-_71040847 | 0.01 |
ENSRNOT00000020606
|
Star
|
steroidogenic acute regulatory protein |
| chr7_+_23854846 | 0.01 |
ENSRNOT00000037290
|
Bpifc
|
BPI fold containing family C |
| chr20_-_32133431 | 0.01 |
ENSRNOT00000000443
|
Srgn
|
serglycin |
| chr1_-_233145924 | 0.01 |
ENSRNOT00000018763
ENSRNOT00000082442 |
Psat1
|
phosphoserine aminotransferase 1 |
| chr5_+_72103704 | 0.01 |
ENSRNOT00000021629
|
Rad23b
|
RAD23 homolog B, nucleotide excision repair protein |
| chr9_+_8052210 | 0.00 |
ENSRNOT00000073659
|
Adgre4
|
adhesion G protein-coupled receptor E4 |
| chr12_+_11240761 | 0.00 |
ENSRNOT00000001310
|
Pdap1
|
PDGFA associated protein 1 |
| chr1_+_89202527 | 0.00 |
ENSRNOT00000028526
|
Sbsn
|
suprabasin |
| chr1_-_16687817 | 0.00 |
ENSRNOT00000091376
ENSRNOT00000081620 |
Myb
|
MYB proto-oncogene, transcription factor |
| chr12_+_38160464 | 0.00 |
ENSRNOT00000032249
|
Hcar2
|
hydroxycarboxylic acid receptor 2 |
| chr8_-_13680870 | 0.00 |
ENSRNOT00000081616
|
Hephl1
|
hephaestin-like 1 |
| chr1_+_22364551 | 0.00 |
ENSRNOT00000043157
|
LOC108348200
|
trace amine-associated receptor 8a |
| chr13_-_82753438 | 0.00 |
ENSRNOT00000075948
|
Atp1b1
|
ATPase Na+/K+ transporting subunit beta 1 |
| chr1_+_276240703 | 0.00 |
ENSRNOT00000022126
|
Acsl5
|
acyl-CoA synthetase long-chain family member 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Cebpd
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0052047 | calcium-independent cell-matrix adhesion(GO:0007161) interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.1 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |