Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
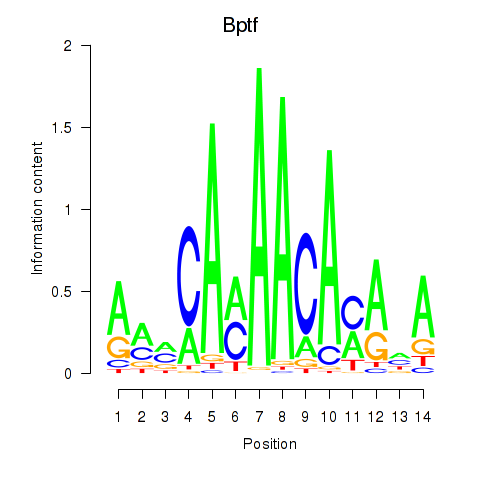
Results for Bptf
Z-value: 0.98
Transcription factors associated with Bptf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Bptf
|
ENSRNOG00000047296 | bromodomain PHD finger transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bptf | rn6_v1_chr10_-_95349789_95349789 | -0.88 | 4.9e-02 | Click! |
Activity profile of Bptf motif
Sorted Z-values of Bptf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_123106694 | 0.94 |
ENSRNOT00000028829
|
Oxt
|
oxytocin/neurophysin 1 prepropeptide |
| chr18_-_15540177 | 0.93 |
ENSRNOT00000022113
|
Ttr
|
transthyretin |
| chr20_+_6356423 | 0.58 |
ENSRNOT00000000628
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr18_-_410098 | 0.57 |
ENSRNOT00000084138
|
LOC102546764
|
cx9C motif-containing protein 4-like |
| chr10_-_27862868 | 0.55 |
ENSRNOT00000004877
|
Gabra6
|
gamma-aminobutyric acid type A receptor alpha 6 subunit |
| chr5_+_172887217 | 0.54 |
ENSRNOT00000022286
|
Tmem52
|
transmembrane protein 52 |
| chr17_+_78793336 | 0.53 |
ENSRNOT00000057898
|
Mt1
|
metallothionein 1 |
| chr2_-_48501436 | 0.51 |
ENSRNOT00000017305
|
Isl1
|
ISL LIM homeobox 1 |
| chr1_-_24302298 | 0.50 |
ENSRNOT00000083452
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr6_-_108415093 | 0.46 |
ENSRNOT00000031650
|
Syndig1l
|
synapse differentiation inducing 1-like |
| chr10_-_56962161 | 0.45 |
ENSRNOT00000026038
|
Alox15
|
arachidonate 15-lipoxygenase |
| chr2_+_236625357 | 0.44 |
ENSRNOT00000081248
|
Papss1
|
3'-phosphoadenosine 5'-phosphosulfate synthase 1 |
| chr5_+_117052260 | 0.42 |
ENSRNOT00000010211
|
Patj
|
PATJ, crumbs cell polarity complex component |
| chr2_-_210116038 | 0.40 |
ENSRNOT00000074950
|
LOC684509
|
similar to NADH-ubiquinone oxidoreductase B9 subunit (Complex I-B9) (CI-B9) |
| chr15_-_60289763 | 0.37 |
ENSRNOT00000038579
|
Fam216b
|
family with sequence similarity 216, member B |
| chr3_-_46726946 | 0.36 |
ENSRNOT00000011030
ENSRNOT00000086576 |
Itgb6
|
integrin subunit beta 6 |
| chrX_-_32153794 | 0.33 |
ENSRNOT00000005348
|
Tmem27
|
transmembrane protein 27 |
| chr10_-_59883839 | 0.32 |
ENSRNOT00000093579
|
Aspa
|
aspartoacylase |
| chr2_-_113616766 | 0.32 |
ENSRNOT00000016858
ENSRNOT00000074723 |
Tmem212
|
transmembrane protein 212 |
| chr3_-_105512939 | 0.32 |
ENSRNOT00000011773
|
Actc1
|
actin, alpha, cardiac muscle 1 |
| chr3_-_147490666 | 0.31 |
ENSRNOT00000046690
|
LOC108351482
|
40S ribosomal protein S29 |
| chr10_+_74724549 | 0.30 |
ENSRNOT00000009077
|
Tex14
|
testis expressed 14, intercellular bridge forming factor |
| chr15_+_51433859 | 0.30 |
ENSRNOT00000093234
|
Tnfrsf10b
|
tumor necrosis factor receptor superfamily, member 10b |
| chr10_+_55626741 | 0.29 |
ENSRNOT00000008492
|
Aurkb
|
aurora kinase B |
| chr8_-_12993651 | 0.29 |
ENSRNOT00000033932
|
Kdm4d
|
lysine demethylase 4D |
| chr5_-_50193571 | 0.28 |
ENSRNOT00000051243
|
Cfap206
|
cilia and flagella associated protein 206 |
| chr5_+_157801163 | 0.27 |
ENSRNOT00000024160
|
Akr7a3
|
aldo-keto reductase family 7 member A3 |
| chr8_+_117906014 | 0.27 |
ENSRNOT00000056180
|
Spink8
|
serine peptidase inhibitor, Kazal type 8 |
| chr1_+_168945449 | 0.26 |
ENSRNOT00000087661
ENSRNOT00000019913 |
LOC103694855
|
hemoglobin subunit beta-2-like |
| chr1_+_168964202 | 0.25 |
ENSRNOT00000089102
|
LOC103694855
|
hemoglobin subunit beta-2-like |
| chr3_-_147819571 | 0.25 |
ENSRNOT00000009840
|
Trib3
|
tribbles pseudokinase 3 |
| chr20_+_32450733 | 0.25 |
ENSRNOT00000036449
|
Rsph4a
|
radial spoke head 4 homolog A |
| chr17_-_61332391 | 0.25 |
ENSRNOT00000034599
|
LOC100362965
|
SNRPN upstream reading frame protein-like |
| chr2_+_210880777 | 0.25 |
ENSRNOT00000026237
|
Gnat2
|
G protein subunit alpha transducin 2 |
| chr18_+_74006046 | 0.25 |
ENSRNOT00000086400
|
AABR07032622.1
|
|
| chr20_+_33945829 | 0.24 |
ENSRNOT00000064063
|
LOC103689994
|
radial spoke head protein 4 homolog A |
| chr10_+_15156207 | 0.24 |
ENSRNOT00000020363
|
Ccdc78
|
coiled-coil domain containing 78 |
| chr5_-_127273656 | 0.23 |
ENSRNOT00000057341
|
Dmrtb1
|
DMRT-like family B with proline-rich C-terminal, 1 |
| chr17_+_43633675 | 0.23 |
ENSRNOT00000072119
|
LOC102549173
|
histone H3.2-like |
| chr5_+_173148884 | 0.23 |
ENSRNOT00000041753
|
LOC100364191
|
hCG1994130-like |
| chr20_-_28873363 | 0.23 |
ENSRNOT00000071064
|
Snrpd2l
|
small nuclear ribonucleoprotein D2-like |
| chr3_-_7498555 | 0.23 |
ENSRNOT00000017725
|
Barhl1
|
BarH-like homeobox 1 |
| chr14_+_87448692 | 0.23 |
ENSRNOT00000077177
|
Igfbp1
|
insulin-like growth factor binding protein 1 |
| chr10_+_59533480 | 0.23 |
ENSRNOT00000087723
|
Atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr1_-_24191908 | 0.23 |
ENSRNOT00000061157
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr15_-_25521393 | 0.22 |
ENSRNOT00000085910
|
Otx2
|
orthodenticle homeobox 2 |
| chr3_+_164002482 | 0.22 |
ENSRNOT00000044141
|
LOC690468
|
similar to 60S ribosomal protein L38 |
| chr10_+_23661013 | 0.22 |
ENSRNOT00000076664
|
Ebf1
|
early B-cell factor 1 |
| chr10_+_23661343 | 0.22 |
ENSRNOT00000047970
|
Ebf1
|
early B-cell factor 1 |
| chr1_-_52962388 | 0.22 |
ENSRNOT00000033685
|
T2
|
brachyury 2 |
| chr8_+_55178289 | 0.22 |
ENSRNOT00000059127
|
Cryab
|
crystallin, alpha B |
| chr20_-_14020007 | 0.22 |
ENSRNOT00000093521
|
Ggt1
|
gamma-glutamyltransferase 1 |
| chr1_+_248132090 | 0.21 |
ENSRNOT00000022056
|
Il33
|
interleukin 33 |
| chr17_+_24654902 | 0.21 |
ENSRNOT00000041960
|
Tfap2a
|
transcription factor AP-2 alpha |
| chr18_-_19275273 | 0.21 |
ENSRNOT00000041707
|
LOC102547344
|
lateral signaling target protein 2 homolog |
| chr6_+_104475036 | 0.21 |
ENSRNOT00000070995
|
Susd6
|
sushi domain containing 6 |
| chr1_-_154111725 | 0.21 |
ENSRNOT00000055488
|
Ccdc81
|
coiled-coil domain containing 81 |
| chr18_+_62852303 | 0.21 |
ENSRNOT00000087673
|
Gnal
|
G protein subunit alpha L |
| chr18_-_59819113 | 0.21 |
ENSRNOT00000065939
|
RGD1562699
|
RGD1562699 |
| chr12_+_44932726 | 0.21 |
ENSRNOT00000085505
|
Rfc5
|
replication factor C subunit 5 |
| chr1_-_91588609 | 0.21 |
ENSRNOT00000050931
|
Snrpg
|
small nuclear ribonucleoprotein polypeptide G |
| chr8_-_43336304 | 0.21 |
ENSRNOT00000036054
|
RGD1311744
|
similar to RIKEN cDNA 5830475I06 |
| chr6_+_93385457 | 0.21 |
ENSRNOT00000010300
|
Actr10
|
actin-related protein 10 homolog |
| chr4_+_33638709 | 0.20 |
ENSRNOT00000009888
ENSRNOT00000034719 ENSRNOT00000052333 |
Tac1
|
tachykinin, precursor 1 |
| chr3_-_111087347 | 0.20 |
ENSRNOT00000018277
|
Rhov
|
ras homolog family member V |
| chr5_+_161889342 | 0.19 |
ENSRNOT00000040481
|
LOC100362684
|
ribosomal protein S20-like |
| chr11_+_67465236 | 0.19 |
ENSRNOT00000042374
|
Stfa2
|
stefin A2 |
| chr13_+_88557860 | 0.19 |
ENSRNOT00000058547
|
Sh2d1b2
|
SH2 domain containing 1B2 |
| chr10_-_66873948 | 0.19 |
ENSRNOT00000039261
|
Evi2a
|
ecotropic viral integration site 2A |
| chr17_+_37675569 | 0.19 |
ENSRNOT00000032995
|
RGD1563300
|
similar to 60S ribosomal protein L29 (P23) |
| chr12_+_48257609 | 0.19 |
ENSRNOT00000031658
ENSRNOT00000090277 |
Alkbh2
|
alkB homolog 2, alpha-ketoglutarate-dependent dioxygenase |
| chr8_-_46694613 | 0.18 |
ENSRNOT00000084514
|
AC133265.1
|
|
| chr6_+_102392828 | 0.18 |
ENSRNOT00000089162
|
Rdh12
|
retinol dehydrogenase 12 (all-trans/9-cis/11-cis) |
| chr4_-_59014416 | 0.18 |
ENSRNOT00000049811
|
RGD1561636
|
similar to 60S ribosomal protein L38 |
| chr2_-_140453107 | 0.18 |
ENSRNOT00000016475
|
Mgarp
|
mitochondria-localized glutamic acid-rich protein |
| chr11_+_64882288 | 0.18 |
ENSRNOT00000077727
|
Pla1a
|
phospholipase A1 member A |
| chr5_-_154438361 | 0.18 |
ENSRNOT00000085003
|
AC141344.2
|
|
| chr17_-_13393243 | 0.18 |
ENSRNOT00000018252
|
Gadd45g
|
growth arrest and DNA-damage-inducible, gamma |
| chrX_-_81866370 | 0.18 |
ENSRNOT00000042570
|
AABR07039651.1
|
|
| chr10_+_64336200 | 0.18 |
ENSRNOT00000046519
|
Rpl37
|
ribosomal protein L37 |
| chr7_-_2623781 | 0.18 |
ENSRNOT00000004173
|
Spryd4
|
SPRY domain containing 4 |
| chr7_+_120901934 | 0.17 |
ENSRNOT00000019323
|
Tomm22
|
translocase of outer mitochondrial membrane 22 |
| chr3_-_113376751 | 0.17 |
ENSRNOT00000030019
|
Catsper2
|
cation channel, sperm associated 2 |
| chr10_+_35680658 | 0.17 |
ENSRNOT00000004111
|
Mrnip
|
MRN complex interacting protein |
| chr10_+_109774878 | 0.17 |
ENSRNOT00000054954
|
Anapc11
|
anaphase promoting complex subunit 11 |
| chr18_-_5314511 | 0.17 |
ENSRNOT00000022637
ENSRNOT00000079682 |
Zfp521
|
zinc finger protein 521 |
| chr12_-_22754491 | 0.17 |
ENSRNOT00000001924
|
Fis1
|
fission, mitochondrial 1 |
| chr2_+_140708397 | 0.17 |
ENSRNOT00000088846
|
Mgst2
|
microsomal glutathione S-transferase 2 |
| chr9_+_46840992 | 0.17 |
ENSRNOT00000019415
|
Il1r2
|
interleukin 1 receptor type 2 |
| chr10_+_68588789 | 0.17 |
ENSRNOT00000049614
|
AABR07030122.1
|
|
| chr16_+_54332660 | 0.17 |
ENSRNOT00000037685
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr5_-_59113773 | 0.17 |
ENSRNOT00000083129
|
AC121204.3
|
|
| chr6_-_146195819 | 0.17 |
ENSRNOT00000007625
|
Sp4
|
Sp4 transcription factor |
| chr10_-_89187474 | 0.17 |
ENSRNOT00000064931
|
Usmg5
|
up-regulated during skeletal muscle growth 5 homolog (mouse) |
| chr10_+_57251253 | 0.17 |
ENSRNOT00000043171
|
LOC108348287
|
60S ribosomal protein L36a |
| chr11_-_71136673 | 0.17 |
ENSRNOT00000042240
|
Fyttd1
|
forty-two-three domain containing 1 |
| chr16_-_6675746 | 0.17 |
ENSRNOT00000025858
|
Prkcd
|
protein kinase C, delta |
| chr3_-_161294125 | 0.17 |
ENSRNOT00000021143
|
Spata25
|
spermatogenesis associated 25 |
| chr12_-_11272997 | 0.17 |
ENSRNOT00000076021
|
Arpc1a
|
actin related protein 2/3 complex, subunit 1A |
| chr1_-_216828581 | 0.17 |
ENSRNOT00000066943
ENSRNOT00000088856 |
Tnfrsf26
|
tumor necrosis factor receptor superfamily, member 26 |
| chr1_-_213811901 | 0.17 |
ENSRNOT00000020265
|
Ifitm3
|
interferon induced transmembrane protein 3 |
| chr1_-_226526959 | 0.17 |
ENSRNOT00000028003
|
Ppp1r32
|
protein phosphatase 1, regulatory subunit 32 |
| chr12_+_8725517 | 0.17 |
ENSRNOT00000001243
|
Slc46a3
|
solute carrier family 46, member 3 |
| chr19_-_53754602 | 0.16 |
ENSRNOT00000035651
|
Fam92b
|
family with sequence similarity 92, member B |
| chr1_+_93242050 | 0.16 |
ENSRNOT00000013741
|
RGD1562402
|
similar to 60S ribosomal protein L27a |
| chr3_-_80052953 | 0.16 |
ENSRNOT00000018998
|
Ddb2
|
damage specific DNA binding protein 2 |
| chr1_+_31967978 | 0.16 |
ENSRNOT00000081471
ENSRNOT00000021532 |
Trip13
|
thyroid hormone receptor interactor 13 |
| chr7_+_124460358 | 0.16 |
ENSRNOT00000014089
|
Tspo
|
translocator protein |
| chr2_+_93792601 | 0.15 |
ENSRNOT00000014701
ENSRNOT00000077311 |
Fabp4
|
fatty acid binding protein 4 |
| chr1_+_7101715 | 0.15 |
ENSRNOT00000019994
|
Sf3b5
|
splicing factor 3b, subunit 5 |
| chr5_-_139933764 | 0.15 |
ENSRNOT00000015278
|
LOC680700
|
similar to ribosomal protein L10a |
| chr8_-_104155775 | 0.15 |
ENSRNOT00000042885
|
LOC102550734
|
60S ribosomal protein L31-like |
| chr13_-_52136127 | 0.15 |
ENSRNOT00000009398
|
Timm17a
|
translocase of inner mitochondrial membrane 17 homolog A (yeast) |
| chr7_+_76980040 | 0.15 |
ENSRNOT00000050726
|
LOC108350501
|
40S ribosomal protein S29 |
| chr15_-_37831031 | 0.15 |
ENSRNOT00000091562
|
Eef1akmt1
|
eukaryotic translation elongation factor 1 alpha lysine methyltransferase 1 |
| chr1_-_214163808 | 0.15 |
ENSRNOT00000082284
|
Rnh1
|
ribonuclease/angiogenin inhibitor 1 |
| chr15_+_17834635 | 0.15 |
ENSRNOT00000085530
|
LOC361016
|
similar to RIKEN cDNA 4933406L09 |
| chr3_-_8315910 | 0.15 |
ENSRNOT00000074482
|
LOC102546892
|
proline-rich protein HaeIII subfamily 1-like |
| chr6_+_22296128 | 0.15 |
ENSRNOT00000087805
|
Dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr1_+_247238798 | 0.15 |
ENSRNOT00000083511
|
Rcl1
|
RNA terminal phosphate cyclase-like 1 |
| chr2_-_3124543 | 0.15 |
ENSRNOT00000036547
|
Fam81b
|
family with sequence similarity 81, member B |
| chr1_+_225037737 | 0.15 |
ENSRNOT00000077959
|
Bscl2
|
BSCL2, seipin lipid droplet biogenesis associated |
| chr2_-_84531192 | 0.14 |
ENSRNOT00000065312
ENSRNOT00000090540 |
Ropn1l
|
rhophilin associated tail protein 1-like |
| chr5_-_167999853 | 0.14 |
ENSRNOT00000087402
|
Park7
|
Parkinsonism associated deglycase |
| chr14_-_85484275 | 0.14 |
ENSRNOT00000083770
|
Kremen1
|
kringle containing transmembrane protein 1 |
| chr1_-_89258935 | 0.14 |
ENSRNOT00000003180
|
LOC100361079
|
ribosomal protein L36-like |
| chr18_-_6587080 | 0.14 |
ENSRNOT00000040815
|
LOC103694404
|
60S ribosomal protein L39 |
| chr2_+_195665454 | 0.14 |
ENSRNOT00000081335
|
Tdrkh
|
tudor and KH domain containing |
| chr3_-_8955538 | 0.14 |
ENSRNOT00000040570
|
LOC686066
|
similar to 60S ribosomal protein L38 |
| chr10_-_63219309 | 0.14 |
ENSRNOT00000076357
|
Blmh
|
bleomycin hydrolase |
| chr10_-_104522241 | 0.14 |
ENSRNOT00000046010
|
Rps18l1
|
ribosomal protein S18-like 1 |
| chr3_-_122703705 | 0.14 |
ENSRNOT00000079923
ENSRNOT00000049857 |
Snrpb
|
small nuclear ribonucleoprotein polypeptides B and B1 |
| chr1_-_277181345 | 0.14 |
ENSRNOT00000038017
ENSRNOT00000038038 |
Nrap
|
nebulin-related anchoring protein |
| chr16_+_2379480 | 0.14 |
ENSRNOT00000079215
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr5_-_144345531 | 0.14 |
ENSRNOT00000014721
|
Tekt2
|
tektin 2 |
| chr10_-_38024937 | 0.14 |
ENSRNOT00000042688
|
Rpl30
|
ribosomal protein L30 |
| chr12_-_7424658 | 0.14 |
ENSRNOT00000050968
|
LOC103690996
|
uncharacterized LOC103690996 |
| chr6_-_111132320 | 0.13 |
ENSRNOT00000016057
|
Ngb
|
neuroglobin |
| chr17_-_11916026 | 0.13 |
ENSRNOT00000046385
|
RGD1561671
|
similar to RIKEN cDNA 2900010M23 |
| chr10_-_46145548 | 0.13 |
ENSRNOT00000033483
|
Pld6
|
phospholipase D family, member 6 |
| chr2_+_164549455 | 0.13 |
ENSRNOT00000017151
|
Mlf1
|
myeloid leukemia factor 1 |
| chr20_+_46707362 | 0.13 |
ENSRNOT00000071045
|
AABR07045411.1
|
|
| chr4_-_152883210 | 0.13 |
ENSRNOT00000056200
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr16_-_21017163 | 0.13 |
ENSRNOT00000027661
|
Mef2b
|
myocyte enhancer factor 2B |
| chr4_-_114823500 | 0.13 |
ENSRNOT00000089870
|
NEWGENE_1562258
|
INO80 complex subunit B |
| chr4_-_99746560 | 0.13 |
ENSRNOT00000012021
|
Mrpl35
|
mitochondrial ribosomal protein L35 |
| chr6_+_108796182 | 0.13 |
ENSRNOT00000006297
|
Fcf1
|
FCF1 rRNA-processing protein |
| chr17_+_15852548 | 0.13 |
ENSRNOT00000022203
|
Card19
|
caspase recruitment domain family, member 19 |
| chr8_+_69753373 | 0.13 |
ENSRNOT00000066973
|
Tipin
|
timeless interacting protein |
| chr10_-_82374171 | 0.12 |
ENSRNOT00000032693
|
Eme1
|
essential meiotic structure-specific endonuclease 1 |
| chr9_+_81535483 | 0.12 |
ENSRNOT00000077285
|
Arpc2
|
actin related protein 2/3 complex, subunit 2 |
| chr15_+_49008958 | 0.12 |
ENSRNOT00000036340
|
AABR07018244.1
|
|
| chr19_+_44164935 | 0.12 |
ENSRNOT00000048998
|
Gabarapl2
|
GABA type A receptor associated protein like 2 |
| chr10_-_15667955 | 0.12 |
ENSRNOT00000085746
ENSRNOT00000027940 |
Mpg
|
N-methylpurine-DNA glycosylase |
| chr17_+_18031228 | 0.12 |
ENSRNOT00000081708
|
Tpmt
|
thiopurine S-methyltransferase |
| chr1_-_281101438 | 0.12 |
ENSRNOT00000012734
|
Rab11fip2
|
RAB11 family interacting protein 2 |
| chr9_-_66019065 | 0.12 |
ENSRNOT00000088729
|
Als2
|
ALS2, alsin Rho guanine nucleotide exchange factor |
| chr5_+_135997052 | 0.12 |
ENSRNOT00000024921
|
Tctex1d4
|
Tctex1 domain containing 4 |
| chr1_-_224921092 | 0.12 |
ENSRNOT00000025196
|
Slc3a2
|
solute carrier family 3 member 2 |
| chr17_-_8980571 | 0.12 |
ENSRNOT00000071426
|
Pcbd2
|
pterin-4 alpha-carbinolamine dehydratase 2 |
| chr10_-_97756521 | 0.12 |
ENSRNOT00000045902
|
Slc16a6
|
solute carrier family 16, member 6 |
| chr9_-_78969013 | 0.12 |
ENSRNOT00000019772
ENSRNOT00000057585 |
Fn1
|
fibronectin 1 |
| chr14_+_104452917 | 0.12 |
ENSRNOT00000050489
|
LOC682793
|
similar to 60S ribosomal protein L38 |
| chr4_+_145580799 | 0.12 |
ENSRNOT00000013727
|
Vhl
|
von Hippel-Lindau tumor suppressor |
| chr17_-_389967 | 0.12 |
ENSRNOT00000023865
|
Fbp2
|
fructose-bisphosphatase 2 |
| chr9_+_69790831 | 0.12 |
ENSRNOT00000045156
|
LOC100360781
|
ribosomal protein L37-like |
| chr3_+_14482388 | 0.12 |
ENSRNOT00000025857
|
Gsn
|
gelsolin |
| chr4_+_100407658 | 0.12 |
ENSRNOT00000018562
|
Capg
|
capping actin protein, gelsolin like |
| chr1_+_114679537 | 0.12 |
ENSRNOT00000019498
|
Oca2
|
OCA2 melanosomal transmembrane protein |
| chr15_+_48789165 | 0.12 |
ENSRNOT00000044562
|
Zfp395
|
zinc finger protein 395 |
| chr2_+_195940715 | 0.12 |
ENSRNOT00000042985
|
LOC100363268
|
rCG31129-like |
| chr20_+_26893016 | 0.12 |
ENSRNOT00000082430
|
Dnajc12
|
DnaJ heat shock protein family (Hsp40) member C12 |
| chr19_+_38643108 | 0.12 |
ENSRNOT00000087392
|
LOC100360619
|
ribosomal protein L28-like |
| chr12_+_12638900 | 0.12 |
ENSRNOT00000001365
ENSRNOT00000080567 |
Ccz1b
|
CCZ1 homolog B, vacuolar protein trafficking and biogenesis associated |
| chr20_+_9743269 | 0.11 |
ENSRNOT00000001533
ENSRNOT00000083505 |
Abcg1
|
ATP binding cassette subfamily G member 1 |
| chr10_+_110631494 | 0.11 |
ENSRNOT00000054915
|
Fn3k
|
fructosamine 3 kinase |
| chr11_-_14160892 | 0.11 |
ENSRNOT00000091864
|
Hspa13
|
heat shock protein 70 family, member 13 |
| chr14_-_33031282 | 0.11 |
ENSRNOT00000043244
|
LOC103693375
|
60S ribosomal protein L39 |
| chr16_-_7412150 | 0.11 |
ENSRNOT00000047252
|
RGD1564325
|
similar to ribosomal protein S24 |
| chr15_+_30413488 | 0.11 |
ENSRNOT00000071781
|
AABR07017733.1
|
|
| chr12_+_47024442 | 0.11 |
ENSRNOT00000001545
|
Cox6a1
|
cytochrome c oxidase subunit 6A1 |
| chr18_-_62965538 | 0.11 |
ENSRNOT00000025164
|
Mppe1
|
metallophosphoesterase 1 |
| chr10_-_10908776 | 0.11 |
ENSRNOT00000065907
|
AABR07072078.1
|
|
| chr9_+_67763897 | 0.11 |
ENSRNOT00000071226
|
Icos
|
inducible T-cell co-stimulator |
| chr1_+_86973745 | 0.11 |
ENSRNOT00000078156
|
Rinl
|
Ras and Rab interactor-like |
| chr14_-_21709084 | 0.11 |
ENSRNOT00000087477
|
Smr3b
|
submaxillary gland androgen regulated protein 3B |
| chr3_+_151335292 | 0.11 |
ENSRNOT00000073642
|
Mmp24
|
matrix metallopeptidase 24 |
| chr20_+_8484311 | 0.11 |
ENSRNOT00000050041
|
LOC100364116
|
ribosomal protein S20-like |
| chr20_-_3372070 | 0.11 |
ENSRNOT00000080770
|
Dhx16
|
DEAH-box helicase 16 |
| chr4_-_93406182 | 0.11 |
ENSRNOT00000007437
|
AABR07060778.1
|
|
| chr2_+_235311719 | 0.11 |
ENSRNOT00000080183
ENSRNOT00000087287 |
Pla2g12a
|
phospholipase A2, group XIIA |
| chr3_+_95939260 | 0.11 |
ENSRNOT00000041291
|
AABR07053188.1
|
|
| chr5_+_167331035 | 0.11 |
ENSRNOT00000024443
|
Rere
|
arginine-glutamic acid dipeptide repeats |
| chr8_-_61595032 | 0.11 |
ENSRNOT00000023428
|
Snx33
|
sorting nexin 33 |
| chr17_-_44744902 | 0.11 |
ENSRNOT00000085381
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr2_+_257911126 | 0.11 |
ENSRNOT00000014725
|
Pigk
|
phosphatidylinositol glycan anchor biosynthesis, class K |
| chr10_+_83104622 | 0.11 |
ENSRNOT00000072972
|
AABR07030375.2
|
|
| chr3_-_24601063 | 0.11 |
ENSRNOT00000037043
|
LOC100362366
|
40S ribosomal protein S17-like |
| chr4_+_172119331 | 0.11 |
ENSRNOT00000010579
|
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr1_+_79754587 | 0.11 |
ENSRNOT00000083211
|
Micb
|
MHC class I polypeptide-related sequence B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Bptf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0043134 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.2 | 0.5 | GO:0048880 | visceral motor neuron differentiation(GO:0021524) sensory system development(GO:0048880) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.1 | 0.5 | GO:0046687 | response to chromate(GO:0046687) |
| 0.1 | 0.3 | GO:0046223 | toxin catabolic process(GO:0009407) mycotoxin metabolic process(GO:0043385) mycotoxin catabolic process(GO:0043387) aflatoxin metabolic process(GO:0046222) aflatoxin catabolic process(GO:0046223) secondary metabolite catabolic process(GO:0090487) organic heteropentacyclic compound metabolic process(GO:1901376) organic heteropentacyclic compound catabolic process(GO:1901377) |
| 0.1 | 0.9 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.4 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 0.5 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.4 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.1 | 0.6 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.1 | 0.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.1 | 0.2 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.1 | 0.7 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 0.4 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.1 | 0.3 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 | 0.2 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.1 | 0.2 | GO:0050787 | enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) TRAIL receptor biosynthetic process(GO:0045557) regulation of TRAIL receptor biosynthetic process(GO:0045560) glycolate biosynthetic process(GO:0046295) detoxification of mercury ion(GO:0050787) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.1 | 0.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.2 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.1 | 0.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.2 | GO:0031179 | peptide modification(GO:0031179) |
| 0.1 | 0.2 | GO:0061517 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) |
| 0.0 | 0.1 | GO:0060816 | random inactivation of X chromosome(GO:0060816) regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.0 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.3 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.0 | 0.1 | GO:0008358 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.0 | 0.2 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.2 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.1 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.0 | 0.3 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.0 | 0.2 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) positive regulation of glucocorticoid secretion(GO:2000851) |
| 0.0 | 0.1 | GO:0015827 | aromatic amino acid transport(GO:0015801) leucine transport(GO:0015820) tryptophan transport(GO:0015827) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:1903923 | protein processing in phagocytic vesicle(GO:1900756) regulation of establishment of T cell polarity(GO:1903903) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.0 | 0.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.3 | GO:1900004 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.3 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.2 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.1 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.0 | 0.2 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.3 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.1 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.1 | GO:2000814 | positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.1 | GO:0090247 | cell motility involved in somitogenic axis elongation(GO:0090247) regulation of cell motility involved in somitogenic axis elongation(GO:0090249) |
| 0.0 | 0.0 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.1 | GO:0046340 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.2 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.1 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.0 | 0.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.1 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.0 | 0.1 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.0 | 0.1 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.2 | GO:0097211 | axonal transport of mitochondrion(GO:0019896) response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.3 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:1901091 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.0 | 0.1 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.0 | GO:0072237 | metanephric proximal tubule development(GO:0072237) |
| 0.0 | 0.0 | GO:0002838 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.1 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.0 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.1 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:1905133 | metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.0 | 0.1 | GO:0015692 | nickel cation transport(GO:0015675) vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) nickel cation transmembrane transport(GO:0035444) |
| 0.0 | 0.1 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.0 | 0.1 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 0.0 | 0.1 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.1 | GO:0033383 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) |
| 0.0 | 0.2 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.0 | GO:0071298 | cellular response to L-ascorbic acid(GO:0071298) |
| 0.0 | 0.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.2 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 0.2 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.2 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.3 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.0 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.0 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.1 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.1 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.1 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.0 | GO:0090172 | microtubule cytoskeleton organization involved in homologous chromosome segregation(GO:0090172) |
| 0.0 | 0.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.0 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 0.1 | GO:1902218 | regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.0 | 0.0 | GO:0060354 | negative regulation of cell adhesion molecule production(GO:0060354) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.0 | GO:0002434 | immune complex clearance(GO:0002434) |
| 0.0 | 0.0 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.0 | GO:0032919 | spermine acetylation(GO:0032919) |
| 0.0 | 0.0 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.2 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.0 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.0 | GO:1905218 | suppression by virus of host apoptotic process(GO:0019050) modulation by virus of host apoptotic process(GO:0039526) cellular process regulating host cell cycle in response to virus(GO:0060154) cellular response to astaxanthin(GO:1905218) |
| 0.0 | 0.0 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.0 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.1 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.0 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.0 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.0 | 0.0 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.0 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.0 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.0 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.0 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) baculum development(GO:1990375) |
| 0.0 | 0.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.5 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.0 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.0 | GO:0003275 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.0 | 0.1 | GO:0007320 | insemination(GO:0007320) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 0.5 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.3 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.3 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.3 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.3 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.2 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.1 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.1 | GO:1990826 | nucleoplasmic periphery of the nuclear pore complex(GO:1990826) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0032301 | recombination nodule(GO:0005713) MutSalpha complex(GO:0032301) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.8 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.0 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.0 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.4 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.3 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 0.4 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.6 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 0.5 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.1 | 0.3 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.1 | 0.7 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.3 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.9 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 0.2 | GO:0036478 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.1 | 0.2 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.2 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.2 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.1 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0034437 | sterol-transporting ATPase activity(GO:0034041) glycoprotein transporter activity(GO:0034437) |
| 0.0 | 1.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.3 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.3 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.3 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.1 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) nickel cation transmembrane transporter activity(GO:0015099) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) chromium ion transmembrane transporter activity(GO:0070835) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.0 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.0 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.0 | 4.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.0 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.3 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.0 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.0 | GO:0052724 | inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.0 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.0 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.0 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.0 | 0.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.6 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |