Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
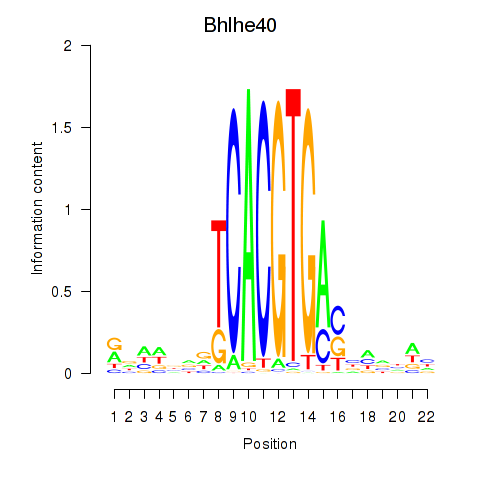
Results for Bhlhe40
Z-value: 0.23
Transcription factors associated with Bhlhe40
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Bhlhe40
|
ENSRNOG00000007152 | basic helix-loop-helix family, member e40 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bhlhe40 | rn6_v1_chr4_+_140703619_140703619 | -0.48 | 4.1e-01 | Click! |
Activity profile of Bhlhe40 motif
Sorted Z-values of Bhlhe40 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_104995725 | 0.07 |
ENSRNOT00000037120
|
Slc25a36
|
solute carrier family 25 member 36 |
| chr10_+_40543288 | 0.06 |
ENSRNOT00000016755
|
Slc36a1
|
solute carrier family 36 member 1 |
| chr1_-_219450451 | 0.06 |
ENSRNOT00000025317
|
Rad9a
|
RAD9 checkpoint clamp component A |
| chr6_-_102472926 | 0.06 |
ENSRNOT00000079351
|
Zfyve26
|
zinc finger FYVE-type containing 26 |
| chrX_-_84821775 | 0.06 |
ENSRNOT00000000174
|
Chm
|
CHM, Rab escort protein 1 |
| chr10_+_94988362 | 0.06 |
ENSRNOT00000066525
|
Cep95
|
centrosomal protein 95 |
| chr1_-_92119951 | 0.05 |
ENSRNOT00000018153
ENSRNOT00000092121 |
Zfp507
|
zinc finger protein 507 |
| chr6_+_109617355 | 0.05 |
ENSRNOT00000011599
|
Flvcr2
|
feline leukemia virus subgroup C cellular receptor family, member 2 |
| chr14_-_44767120 | 0.05 |
ENSRNOT00000003991
|
Wdr19
|
WD repeat domain 19 |
| chr9_+_94425252 | 0.05 |
ENSRNOT00000064965
ENSRNOT00000076099 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr18_+_15298978 | 0.05 |
ENSRNOT00000021263
|
Trappc8
|
trafficking protein particle complex 8 |
| chr10_-_62287189 | 0.05 |
ENSRNOT00000004365
|
Wdr81
|
WD repeat domain 81 |
| chr5_-_160352927 | 0.04 |
ENSRNOT00000017247
|
Dnajc16
|
DnaJ heat shock protein family (Hsp40) member C16 |
| chr1_-_144601327 | 0.04 |
ENSRNOT00000029244
ENSRNOT00000078144 |
Saxo2
|
stabilizer of axonemal microtubules 2 |
| chr7_+_53878610 | 0.04 |
ENSRNOT00000091910
|
Osbpl8
|
oxysterol binding protein-like 8 |
| chr10_-_6870011 | 0.04 |
ENSRNOT00000003439
|
RGD1309748
|
similar to CG4768-PA |
| chr3_+_163570532 | 0.04 |
ENSRNOT00000010054
|
Arfgef2
|
ADP ribosylation factor guanine nucleotide exchange factor 2 |
| chr2_-_210088949 | 0.04 |
ENSRNOT00000070994
|
Rbm15
|
RNA binding motif protein 15 |
| chr10_+_62191656 | 0.04 |
ENSRNOT00000029866
|
Smyd4
|
SET and MYND domain containing 4 |
| chr18_+_55797198 | 0.04 |
ENSRNOT00000026334
ENSRNOT00000026394 |
Dctn4
|
dynactin subunit 4 |
| chr1_+_154377447 | 0.03 |
ENSRNOT00000084268
ENSRNOT00000092086 ENSRNOT00000091470 ENSRNOT00000025415 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr12_+_38459832 | 0.02 |
ENSRNOT00000090343
|
Vps33a
|
VPS33A CORVET/HOPS core subunit |
| chr10_-_94988461 | 0.02 |
ENSRNOT00000048490
|
Ddx5
|
DEAD-box helicase 5 |
| chr5_+_138470069 | 0.02 |
ENSRNOT00000076343
ENSRNOT00000064073 |
Zmynd12
|
zinc finger, MYND-type containing 12 |
| chr2_-_196270826 | 0.02 |
ENSRNOT00000028609
|
Pip5k1a
|
phosphatidylinositol-4-phosphate 5-kinase, type 1, alpha |
| chrX_+_74200972 | 0.02 |
ENSRNOT00000076956
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr19_+_14392423 | 0.02 |
ENSRNOT00000018880
|
Tom1
|
target of myb1 membrane trafficking protein |
| chr19_+_26151900 | 0.02 |
ENSRNOT00000005571
|
Tnpo2
|
transportin 2 |
| chrX_-_105417323 | 0.02 |
ENSRNOT00000015494
|
Gla
|
galactosidase, alpha |
| chr1_-_222495382 | 0.02 |
ENSRNOT00000028759
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr6_-_122239614 | 0.02 |
ENSRNOT00000005015
|
Galc
|
galactosylceramidase |
| chr1_+_144601410 | 0.02 |
ENSRNOT00000047408
|
Efl1
|
elongation factor like GTPase 1 |
| chr4_+_148139528 | 0.02 |
ENSRNOT00000092594
ENSRNOT00000015620 ENSRNOT00000092613 |
Washc2c
|
WASH complex subunit 2C |
| chr10_-_62191512 | 0.01 |
ENSRNOT00000090262
|
Rpa1
|
replication protein A1 |
| chr10_-_10725655 | 0.01 |
ENSRNOT00000061236
|
Ubn1
|
ubinuclein 1 |
| chr13_-_70174565 | 0.01 |
ENSRNOT00000067135
|
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr10_+_10725819 | 0.01 |
ENSRNOT00000004159
|
Glyr1
|
glyoxylate reductase 1 homolog |
| chr1_-_164101578 | 0.01 |
ENSRNOT00000022176
|
Uvrag
|
UV radiation resistance associated |
| chr13_-_111765944 | 0.01 |
ENSRNOT00000073041
|
Syt14
|
synaptotagmin 14 |
| chr10_-_37455022 | 0.01 |
ENSRNOT00000016742
|
LOC103694902
|
ubiquitin-conjugating enzyme E2 B |
| chr1_+_200167169 | 0.01 |
ENSRNOT00000027665
|
Sec23ip
|
SEC23 interacting protein |
| chr2_+_198852500 | 0.01 |
ENSRNOT00000000110
|
Rnf115
|
ring finger protein 115 |
| chr1_-_102849430 | 0.01 |
ENSRNOT00000086856
|
Saa4
|
serum amyloid A4 |
| chr16_+_8497569 | 0.01 |
ENSRNOT00000027054
|
Ogdhl
|
oxoglutarate dehydrogenase-like |
| chr20_-_32115310 | 0.01 |
ENSRNOT00000067883
ENSRNOT00000081916 |
Vps26a
|
VPS26 retromer complex component A |
| chr2_-_198852161 | 0.01 |
ENSRNOT00000028815
|
Polr3c
|
RNA polymerase III subunit C |
| chr1_-_72329856 | 0.01 |
ENSRNOT00000021391
|
U2af2
|
U2 small nuclear RNA auxiliary factor 2 |
| chr8_+_81863619 | 0.01 |
ENSRNOT00000080608
|
Fam214a
|
family with sequence similarity 214, member A |
| chr11_+_60613882 | 0.01 |
ENSRNOT00000002853
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr5_-_164844586 | 0.00 |
ENSRNOT00000011287
|
Clcn6
|
chloride voltage-gated channel 6 |
| chr6_-_12997817 | 0.00 |
ENSRNOT00000022802
|
Fshr
|
follicle stimulating hormone receptor |
| chr19_-_43215077 | 0.00 |
ENSRNOT00000082151
|
Aars
|
alanyl-tRNA synthetase |
| chr3_+_138504214 | 0.00 |
ENSRNOT00000091529
|
Kat14
|
lysine acetyltransferase 14 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Bhlhe40
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) myotome development(GO:0061055) |
| 0.0 | 0.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.0 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |