Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
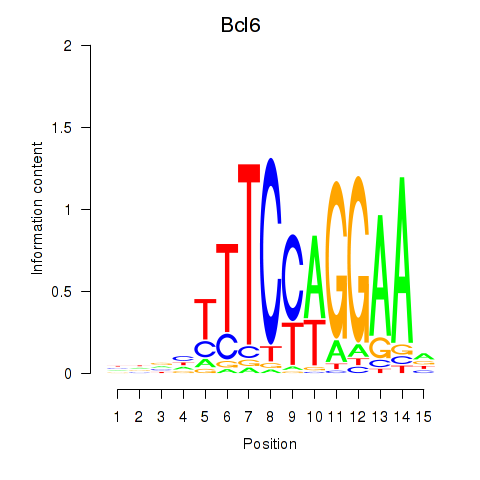
Results for Bcl6
Z-value: 0.47
Transcription factors associated with Bcl6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Bcl6
|
ENSRNOG00000001843 | B-cell CLL/lymphoma 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bcl6 | rn6_v1_chr11_+_80255790_80255790 | 0.89 | 4.2e-02 | Click! |
Activity profile of Bcl6 motif
Sorted Z-values of Bcl6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_3342491 | 0.34 |
ENSRNOT00000081756
|
Rdh5
|
retinol dehydrogenase 5 |
| chr3_-_2803574 | 0.24 |
ENSRNOT00000040995
|
RGD1560470
|
similar to Gene model 996 |
| chr5_+_29538380 | 0.23 |
ENSRNOT00000010845
|
Calb1
|
calbindin 1 |
| chr7_+_18510354 | 0.23 |
ENSRNOT00000060002
|
Pram1
|
PML-RARA regulated adaptor molecule 1 |
| chr16_+_54291251 | 0.22 |
ENSRNOT00000079006
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr19_-_37528011 | 0.19 |
ENSRNOT00000059628
|
Agrp
|
agouti related neuropeptide |
| chr5_+_139597731 | 0.19 |
ENSRNOT00000072427
|
Cited4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr8_-_80631873 | 0.16 |
ENSRNOT00000091661
ENSRNOT00000080662 |
Unc13c
|
unc-13 homolog C |
| chr2_-_259150479 | 0.16 |
ENSRNOT00000085892
|
AABR07013843.1
|
|
| chr16_+_54358471 | 0.15 |
ENSRNOT00000047314
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr10_-_61361250 | 0.14 |
ENSRNOT00000092203
|
Rap1gap2
|
RAP1 GTPase activating protein 2 |
| chr13_+_90723092 | 0.14 |
ENSRNOT00000010146
|
Kcnj10
|
ATP-sensitive inward rectifier potassium channel 10 |
| chr5_-_59638385 | 0.14 |
ENSRNOT00000060203
ENSRNOT00000077367 |
Rnf38
|
ring finger protein 38 |
| chr5_-_153625869 | 0.13 |
ENSRNOT00000024464
|
Clic4
|
chloride intracellular channel 4 |
| chr12_-_10307265 | 0.13 |
ENSRNOT00000092627
|
Wasf3
|
WAS protein family, member 3 |
| chr8_-_12902262 | 0.12 |
ENSRNOT00000033969
|
Endod1
|
endonuclease domain containing 1 |
| chr10_+_105393072 | 0.12 |
ENSRNOT00000013359
|
Ubald2
|
UBA-like domain containing 2 |
| chr19_-_57333433 | 0.12 |
ENSRNOT00000024917
|
Agt
|
angiotensinogen |
| chr4_+_9981958 | 0.11 |
ENSRNOT00000015322
|
Napepld
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr4_-_123494742 | 0.11 |
ENSRNOT00000073268
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr17_-_10818835 | 0.11 |
ENSRNOT00000091046
|
Cplx2
|
complexin 2 |
| chr17_+_78739389 | 0.11 |
ENSRNOT00000020384
|
LOC102547897
|
uncharacterized LOC102547897 |
| chr12_+_38965063 | 0.11 |
ENSRNOT00000074936
|
Morn3
|
MORN repeat containing 3 |
| chr4_-_30556814 | 0.11 |
ENSRNOT00000012760
|
Pdk4
|
pyruvate dehydrogenase kinase 4 |
| chr4_+_146455332 | 0.10 |
ENSRNOT00000009775
|
Hrh1
|
histamine receptor H 1 |
| chr3_+_177310753 | 0.10 |
ENSRNOT00000031448
|
Myt1
|
myelin transcription factor 1 |
| chr10_-_59883839 | 0.10 |
ENSRNOT00000093579
|
Aspa
|
aspartoacylase |
| chr4_-_117575154 | 0.10 |
ENSRNOT00000075813
|
LOC102556148
|
probable N-acetyltransferase CML2-like |
| chr13_-_45068077 | 0.10 |
ENSRNOT00000004969
|
Mcm6
|
minichromosome maintenance complex component 6 |
| chr19_-_25801526 | 0.10 |
ENSRNOT00000003884
|
Nacc1
|
nucleus accumbens associated 1 |
| chr7_-_15163866 | 0.09 |
ENSRNOT00000042638
|
Zfp870
|
zinc finger protein 870 |
| chr5_-_2982603 | 0.09 |
ENSRNOT00000045460
|
Kcnb2
|
potassium voltage-gated channel subfamily B member 2 |
| chr4_+_163162211 | 0.09 |
ENSRNOT00000082537
|
Clec1b
|
C-type lectin domain family 1, member B |
| chr3_-_125213607 | 0.09 |
ENSRNOT00000078070
|
Gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| chr2_-_200003443 | 0.09 |
ENSRNOT00000024900
ENSRNOT00000088041 |
Pde4dip
|
phosphodiesterase 4D interacting protein |
| chr8_+_116054465 | 0.08 |
ENSRNOT00000040056
|
Cish
|
cytokine inducible SH2-containing protein |
| chr5_+_169452493 | 0.08 |
ENSRNOT00000014444
|
Gpr153
|
G protein-coupled receptor 153 |
| chr17_-_14557792 | 0.08 |
ENSRNOT00000078166
|
LOC103690033
|
isoleucine--tRNA ligase, cytoplasmic-like |
| chr16_-_81945127 | 0.08 |
ENSRNOT00000023352
|
Mcf2l
|
MCF.2 cell line derived transforming sequence-like |
| chr9_-_17209220 | 0.08 |
ENSRNOT00000083764
|
Gtpbp2
|
GTP binding protein 2 |
| chr5_+_164845925 | 0.08 |
ENSRNOT00000011384
|
Mthfr
|
methylenetetrahydrofolate reductase |
| chr12_-_11780078 | 0.08 |
ENSRNOT00000076796
|
Zfp498
|
zinc finger protein 498 |
| chr4_+_154423209 | 0.08 |
ENSRNOT00000075799
|
LOC100911545
|
alpha-2-macroglobulin-like |
| chrX_+_28072826 | 0.08 |
ENSRNOT00000039796
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr2_+_104744461 | 0.08 |
ENSRNOT00000016083
ENSRNOT00000082627 |
Cp
|
ceruloplasmin |
| chr17_-_43627629 | 0.08 |
ENSRNOT00000022965
|
Hist1h2af
|
histone cluster 1, H2af |
| chr1_-_250774105 | 0.08 |
ENSRNOT00000079227
|
Sgms1
|
sphingomyelin synthase 1 |
| chr2_-_165600748 | 0.07 |
ENSRNOT00000013216
|
Ift80
|
intraflagellar transport 80 |
| chr1_+_190666149 | 0.07 |
ENSRNOT00000089361
|
LOC102547219
|
uncharacterized LOC102547219 |
| chr5_-_164585634 | 0.07 |
ENSRNOT00000065985
|
Mfn2
|
mitofusin 2 |
| chr5_+_58855773 | 0.06 |
ENSRNOT00000072869
|
Rusc2
|
RUN and SH3 domain containing 2 |
| chr10_+_58784660 | 0.06 |
ENSRNOT00000019925
|
LOC100912483
|
uncharacterized LOC100912483 |
| chr1_+_106998623 | 0.06 |
ENSRNOT00000022383
|
Slc17a6
|
solute carrier family 17 member 6 |
| chr17_-_43821536 | 0.06 |
ENSRNOT00000072286
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr1_+_8310577 | 0.06 |
ENSRNOT00000015131
|
Hivep2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr5_+_22392732 | 0.06 |
ENSRNOT00000087182
|
Clvs1
|
clavesin 1 |
| chr1_+_156552328 | 0.06 |
ENSRNOT00000055401
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr8_-_130429132 | 0.06 |
ENSRNOT00000026261
|
Hhatl
|
hedgehog acyltransferase-like |
| chr4_+_55772627 | 0.06 |
ENSRNOT00000048252
|
Snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr2_+_53109684 | 0.06 |
ENSRNOT00000086590
|
Selenop
|
selenoprotein P |
| chr7_+_42269784 | 0.06 |
ENSRNOT00000008471
ENSRNOT00000007231 |
Kitlg
|
KIT ligand |
| chr3_-_117990289 | 0.06 |
ENSRNOT00000011084
|
Shc4
|
SHC adaptor protein 4 |
| chr15_-_33285779 | 0.06 |
ENSRNOT00000036150
|
Cdh24
|
cadherin 24 |
| chr20_+_27954433 | 0.06 |
ENSRNOT00000064288
|
Lims1
|
LIM zinc finger domain containing 1 |
| chr4_+_154309426 | 0.06 |
ENSRNOT00000019346
|
A2m
|
alpha-2-macroglobulin |
| chrX_+_71199491 | 0.06 |
ENSRNOT00000076168
ENSRNOT00000005077 ENSRNOT00000005102 |
Nlgn3
|
neuroligin 3 |
| chr10_+_63803309 | 0.06 |
ENSRNOT00000036666
|
Myo1c
|
myosin 1C |
| chr7_-_139223116 | 0.06 |
ENSRNOT00000086559
|
Endou
|
endonuclease, poly(U)-specific |
| chr15_+_23792931 | 0.06 |
ENSRNOT00000092091
|
Samd4a
|
sterile alpha motif domain containing 4A |
| chr12_+_16030788 | 0.06 |
ENSRNOT00000051565
|
Iqce
|
IQ motif containing E |
| chr18_+_48853418 | 0.05 |
ENSRNOT00000022792
|
Csnk1g3
|
casein kinase 1, gamma 3 |
| chr16_-_19996716 | 0.05 |
ENSRNOT00000024434
ENSRNOT00000080706 |
Nxnl1
|
nucleoredoxin-like 1 |
| chr5_+_78428669 | 0.05 |
ENSRNOT00000080744
|
Rgs3
|
regulator of G-protein signaling 3 |
| chr7_-_94563001 | 0.05 |
ENSRNOT00000051139
ENSRNOT00000005561 |
Enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr2_-_140334912 | 0.05 |
ENSRNOT00000015067
|
Elf2
|
E74-like factor 2 |
| chr1_+_31124825 | 0.05 |
ENSRNOT00000092105
|
AABR07000989.1
|
|
| chrX_+_22313028 | 0.05 |
ENSRNOT00000082725
|
Kdm5c
|
lysine demethylase 5C |
| chr16_+_74076812 | 0.05 |
ENSRNOT00000025675
|
Ap3m2
|
adaptor-related protein complex 3, mu 2 subunit |
| chr1_-_101697277 | 0.05 |
ENSRNOT00000028549
|
Sphk2
|
sphingosine kinase 2 |
| chr9_+_51302151 | 0.05 |
ENSRNOT00000085908
|
Gulp1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr4_+_147037179 | 0.05 |
ENSRNOT00000011292
|
Syn2
|
synapsin II |
| chr8_+_22750336 | 0.05 |
ENSRNOT00000013496
|
Ldlr
|
low density lipoprotein receptor |
| chr12_-_38916237 | 0.05 |
ENSRNOT00000074517
|
Tmem120b
|
transmembrane protein 120B |
| chr1_+_221710670 | 0.05 |
ENSRNOT00000064798
|
Map4k2
|
mitogen activated protein kinase kinase kinase kinase 2 |
| chr1_-_188097374 | 0.05 |
ENSRNOT00000092246
|
Syt17
|
synaptotagmin 17 |
| chr4_-_10329241 | 0.05 |
ENSRNOT00000017232
|
Fgl2
|
fibrinogen-like 2 |
| chr12_+_9728486 | 0.05 |
ENSRNOT00000001263
|
Lnx2
|
ligand of numb-protein X 2 |
| chr4_-_11610518 | 0.04 |
ENSRNOT00000066643
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr2_+_78245459 | 0.04 |
ENSRNOT00000089551
|
LOC103689968
|
protein FAM134B |
| chr14_+_8182383 | 0.04 |
ENSRNOT00000092719
|
Mapk10
|
mitogen activated protein kinase 10 |
| chrX_-_123092217 | 0.04 |
ENSRNOT00000039710
|
Gm14569
|
predicted gene 14569 |
| chr14_+_115275894 | 0.04 |
ENSRNOT00000033437
|
Gpr75
|
G protein-coupled receptor 75 |
| chr12_-_38995570 | 0.04 |
ENSRNOT00000001806
|
Orai1
|
ORAI calcium release-activated calcium modulator 1 |
| chr2_+_144646308 | 0.04 |
ENSRNOT00000078337
ENSRNOT00000093407 |
Dclk1
|
doublecortin-like kinase 1 |
| chr4_+_67059939 | 0.04 |
ENSRNOT00000052393
|
Chmp4bl1
|
chromatin modifying protein 4B-like 1 |
| chr15_-_34392066 | 0.04 |
ENSRNOT00000027315
|
Tgm1
|
transglutaminase 1 |
| chr6_-_105160470 | 0.04 |
ENSRNOT00000008367
|
Adam4
|
a disintegrin and metalloprotease domain 4 |
| chr10_-_45579029 | 0.04 |
ENSRNOT00000080028
|
Arf1
|
ADP-ribosylation factor 1 |
| chr4_-_77747070 | 0.04 |
ENSRNOT00000009247
|
Zfp746
|
zinc finger protein 746 |
| chr2_+_60920257 | 0.04 |
ENSRNOT00000025170
|
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr14_-_37294472 | 0.04 |
ENSRNOT00000081087
|
Dcun1d4
|
defective in cullin neddylation 1 domain containing 4 |
| chr3_-_161376119 | 0.04 |
ENSRNOT00000023521
|
Zfp335
|
zinc finger protein 335 |
| chr20_-_6251634 | 0.04 |
ENSRNOT00000059194
|
Stk38
|
serine/threonine kinase 38 |
| chr3_-_2060167 | 0.04 |
ENSRNOT00000066777
|
Ehmt1
|
euchromatic histone lysine methyltransferase 1 |
| chr9_-_38196273 | 0.04 |
ENSRNOT00000044452
|
Dst
|
dystonin |
| chr9_+_100104000 | 0.04 |
ENSRNOT00000074160
|
Capn10
|
calpain 10 |
| chr1_-_77844189 | 0.04 |
ENSRNOT00000017555
|
Gltscr2
|
glioma tumor suppressor candidate region gene 2 |
| chr7_-_144109116 | 0.04 |
ENSRNOT00000020437
|
Map3k12
|
mitogen activated protein kinase kinase kinase 12 |
| chr17_+_36691875 | 0.04 |
ENSRNOT00000074314
|
LOC688583
|
similar to High mobility group protein 4 (HMG-4) (High mobility group protein 2a) (HMG-2a) |
| chr1_+_85112834 | 0.04 |
ENSRNOT00000026107
|
Dyrk1b
|
dual specificity tyrosine phosphorylation regulated kinase 1B |
| chr8_+_114300257 | 0.03 |
ENSRNOT00000090707
ENSRNOT00000018654 |
Pik3r4
|
phosphoinositide-3-kinase, regulatory subunit 4 |
| chr1_-_89360733 | 0.03 |
ENSRNOT00000028544
|
Mag
|
myelin-associated glycoprotein |
| chr3_-_111873974 | 0.03 |
ENSRNOT00000077546
|
AABR07053509.2
|
|
| chr13_+_84474319 | 0.03 |
ENSRNOT00000031367
ENSRNOT00000072244 ENSRNOT00000072897 ENSRNOT00000064168 ENSRNOT00000074954 ENSRNOT00000073696 |
Ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr11_-_65209268 | 0.03 |
ENSRNOT00000077612
|
Gsk3b
|
glycogen synthase kinase 3 beta |
| chr16_-_19097329 | 0.03 |
ENSRNOT00000030617
|
RGD1311847
|
similar to 1700030K09Rik protein |
| chr1_-_98493978 | 0.03 |
ENSRNOT00000023942
|
Nkg7
|
natural killer cell granule protein 7 |
| chr11_-_86357718 | 0.03 |
ENSRNOT00000071806
|
Cldn5
|
claudin 5 |
| chr7_-_122329443 | 0.03 |
ENSRNOT00000032232
|
Mkl1
|
megakaryoblastic leukemia (translocation) 1 |
| chr9_-_61134963 | 0.03 |
ENSRNOT00000017967
|
Pgap1
|
post-GPI attachment to proteins 1 |
| chr15_+_103344476 | 0.03 |
ENSRNOT00000013329
ENSRNOT00000077047 ENSRNOT00000078103 |
Gpr180
|
G protein-coupled receptor 180 |
| chr19_+_43220508 | 0.03 |
ENSRNOT00000091788
ENSRNOT00000084462 ENSRNOT00000087474 |
Ddx19a
|
DEAD-box helicase 19A |
| chrX_+_20423401 | 0.02 |
ENSRNOT00000093162
|
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr8_+_21890319 | 0.02 |
ENSRNOT00000027949
|
LOC500956
|
hypothetical protein LOC500956 |
| chr14_-_42560174 | 0.02 |
ENSRNOT00000003128
|
Tmem33
|
transmembrane protein 33 |
| chr7_+_139685573 | 0.02 |
ENSRNOT00000088376
|
Pfkm
|
phosphofructokinase, muscle |
| chr2_-_52282548 | 0.02 |
ENSRNOT00000033627
|
Nnt
|
nicotinamide nucleotide transhydrogenase |
| chr3_-_112084144 | 0.02 |
ENSRNOT00000010959
|
Pla2g4f
|
phospholipase A2, group IVF |
| chr18_+_17550350 | 0.02 |
ENSRNOT00000078870
|
RGD1562608
|
similar to KIAA1328 protein |
| chr6_+_109110534 | 0.02 |
ENSRNOT00000009156
|
Zc2hc1c
|
zinc finger, C2HC-type containing 1C |
| chr2_+_58448917 | 0.02 |
ENSRNOT00000082562
|
Ranbp3l
|
RAN binding protein 3-like |
| chr16_-_69961162 | 0.02 |
ENSRNOT00000074955
|
Prrg3
|
proline rich and Gla domain 3 |
| chr16_+_75145401 | 0.02 |
ENSRNOT00000046821
ENSRNOT00000047761 |
Spag11bl
|
sperm associated antigen 11b-like |
| chr12_-_39769480 | 0.02 |
ENSRNOT00000036137
|
Pptc7
|
PTC7 protein phosphatase homolog |
| chr1_+_225037737 | 0.02 |
ENSRNOT00000077959
|
Bscl2
|
BSCL2, seipin lipid droplet biogenesis associated |
| chr5_+_79383511 | 0.02 |
ENSRNOT00000011799
|
Tmem268
|
transmembrane protein 268 |
| chr16_+_23553647 | 0.02 |
ENSRNOT00000041994
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr20_+_5933303 | 0.02 |
ENSRNOT00000000617
|
Mapk14
|
mitogen activated protein kinase 14 |
| chr18_+_56364620 | 0.02 |
ENSRNOT00000068535
ENSRNOT00000086033 |
Pdgfrb
|
platelet derived growth factor receptor beta |
| chr10_+_90342051 | 0.02 |
ENSRNOT00000028487
|
Rundc3a
|
RUN domain containing 3A |
| chr8_-_36764422 | 0.02 |
ENSRNOT00000017330
|
Hyls1
|
HYLS1, centriolar and ciliogenesis associated |
| chr9_+_47987 | 0.02 |
ENSRNOT00000066582
|
Efhb
|
EF hand domain family, member B |
| chr6_+_95323579 | 0.01 |
ENSRNOT00000007369
|
Pcnx4
|
pecanex homolog 4 (Drosophila) |
| chr17_+_53231343 | 0.01 |
ENSRNOT00000021703
|
Hecw1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr17_+_44025267 | 0.01 |
ENSRNOT00000023268
|
Zfp322a
|
zinc finger protein 322a |
| chr4_-_58195143 | 0.01 |
ENSRNOT00000033706
|
Copg2
|
coatomer protein complex, subunit gamma 2 |
| chr8_-_48634797 | 0.01 |
ENSRNOT00000012868
|
Hinfp
|
histone H4 transcription factor |
| chr9_+_10952374 | 0.01 |
ENSRNOT00000074993
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr4_-_168656673 | 0.01 |
ENSRNOT00000009341
|
Gpr19
|
G protein-coupled receptor 19 |
| chr1_+_220362064 | 0.01 |
ENSRNOT00000074361
|
LOC108348067
|
endosialin |
| chr10_-_89088993 | 0.01 |
ENSRNOT00000027458
|
Ccr10
|
C-C motif chemokine receptor 10 |
| chr12_-_41266430 | 0.01 |
ENSRNOT00000001853
ENSRNOT00000081108 |
Oas1b
|
2-5 oligoadenylate synthetase 1B |
| chr6_+_96871625 | 0.01 |
ENSRNOT00000012361
|
Snapc1
|
small nuclear RNA activating complex, polypeptide 1 |
| chr10_-_84789832 | 0.01 |
ENSRNOT00000071719
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr8_-_32000378 | 0.01 |
ENSRNOT00000013341
|
Adamts15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15 |
| chr12_+_46869836 | 0.01 |
ENSRNOT00000084421
|
Sirt4
|
sirtuin 4 |
| chrX_+_9436707 | 0.01 |
ENSRNOT00000004187
|
Cask
|
calcium/calmodulin dependent serine protein kinase |
| chr10_-_91699424 | 0.01 |
ENSRNOT00000004650
|
Lyzl6
|
lysozyme-like 6 |
| chr5_-_113939127 | 0.01 |
ENSRNOT00000039554
|
Mysm1
|
myb-like, SWIRM and MPN domains 1 |
| chr1_+_260093641 | 0.01 |
ENSRNOT00000019521
|
Ccnj
|
cyclin J |
| chr15_+_55461695 | 0.01 |
ENSRNOT00000065195
|
Sucla2
|
succinate-CoA ligase ADP-forming beta subunit |
| chr1_-_64162461 | 0.01 |
ENSRNOT00000091739
|
Prpf31
|
pre-mRNA processing factor 31 |
| chr14_+_21972274 | 0.01 |
ENSRNOT00000060265
|
Csn2
|
casein beta |
| chr4_-_150829913 | 0.01 |
ENSRNOT00000041571
|
Cacna1c
|
calcium voltage-gated channel subunit alpha1 C |
| chr5_+_156587093 | 0.01 |
ENSRNOT00000019696
|
Hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr1_-_200696928 | 0.01 |
ENSRNOT00000068183
ENSRNOT00000022253 ENSRNOT00000065448 ENSRNOT00000022331 |
Fgfr2
|
fibroblast growth factor receptor 2 |
| chr3_-_140728129 | 0.00 |
ENSRNOT00000015637
|
Ralgapa2
|
Ral GTPase activating protein catalytic alpha subunit 2 |
| chr12_-_48218955 | 0.00 |
ENSRNOT00000067975
ENSRNOT00000080557 ENSRNOT00000000821 |
Acacb
|
acetyl-CoA carboxylase beta |
| chr8_-_126390801 | 0.00 |
ENSRNOT00000089732
|
AABR07071661.1
|
|
| chr3_+_56862691 | 0.00 |
ENSRNOT00000087712
|
Gad1
|
glutamate decarboxylase 1 |
| chr1_+_228317412 | 0.00 |
ENSRNOT00000028575
|
Olr319
|
olfactory receptor 319 |
| chr12_-_46493203 | 0.00 |
ENSRNOT00000057036
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chr4_-_55398941 | 0.00 |
ENSRNOT00000075324
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr9_+_44681494 | 0.00 |
ENSRNOT00000035785
|
Eif5b
|
eukaryotic translation initiation factor 5B |
| chr8_+_85503224 | 0.00 |
ENSRNOT00000012348
|
Gsta4
|
glutathione S-transferase alpha 4 |
| chr9_+_10995198 | 0.00 |
ENSRNOT00000089071
|
Ubxn6
|
UBX domain protein 6 |
| chr16_-_71237118 | 0.00 |
ENSRNOT00000066974
|
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr11_-_70207361 | 0.00 |
ENSRNOT00000002444
|
Muc13
|
mucin 13, cell surface associated |
| chrX_-_70835089 | 0.00 |
ENSRNOT00000076351
|
Tex11
|
testis expressed 11 |
| chr6_-_107080524 | 0.00 |
ENSRNOT00000011662
|
Zfyve1
|
zinc finger FYVE-type containing 1 |
| chr20_+_2194709 | 0.00 |
ENSRNOT00000001017
|
Trim15
|
tripartite motif containing 15 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Bcl6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.0 | 0.1 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 | 0.1 | GO:0003331 | ovarian follicle rupture(GO:0001543) angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) regulation of renal output by angiotensin(GO:0002019) angiotensin-mediated drinking behavior(GO:0003051) regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) response to muscle activity involved in regulation of muscle adaptation(GO:0014873) positive regulation of gap junction assembly(GO:1903598) |
| 0.0 | 0.2 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.0 | 0.1 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.1 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.0 | 0.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:1903165 | response to polycyclic arene(GO:1903165) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.1 | GO:0070666 | negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.0 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 0.1 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.0 | 0.0 | GO:0010899 | regulation of phosphatidylcholine catabolic process(GO:0010899) receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.0 | 0.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.0 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.1 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.0 | GO:0097453 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0099510 | calcium ion binding involved in regulation of cytosolic calcium ion concentration(GO:0099510) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.0 | GO:0070699 | type II activin receptor binding(GO:0070699) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |