Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
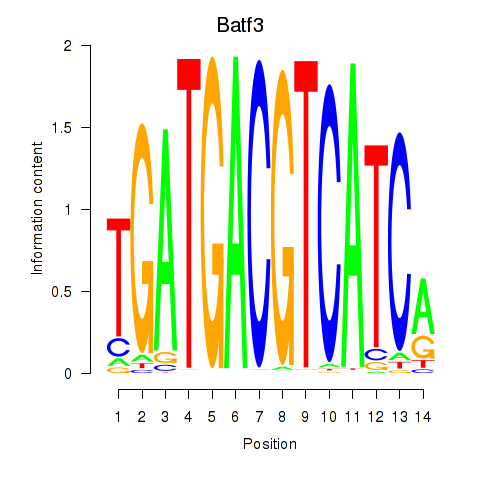
Results for Batf3
Z-value: 0.36
Transcription factors associated with Batf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Batf3
|
ENSRNOG00000003716 | basic leucine zipper ATF-like transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Batf3 | rn6_v1_chr13_+_109713489_109713489 | -0.08 | 9.0e-01 | Click! |
Activity profile of Batf3 motif
Sorted Z-values of Batf3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_114866768 | 0.36 |
ENSRNOT00000076731
|
Wdr82
|
WD repeat domain 82 |
| chr1_+_218466289 | 0.19 |
ENSRNOT00000017948
|
Mrgprf
|
MAS related GPR family member F |
| chr4_+_148139528 | 0.19 |
ENSRNOT00000092594
ENSRNOT00000015620 ENSRNOT00000092613 |
Washc2c
|
WASH complex subunit 2C |
| chrX_-_45522665 | 0.17 |
ENSRNOT00000030771
|
RGD1562200
|
similar to GS2 gene |
| chr7_+_120580743 | 0.08 |
ENSRNOT00000017181
|
Maff
|
MAF bZIP transcription factor F |
| chr6_+_28515025 | 0.08 |
ENSRNOT00000088033
ENSRNOT00000005317 ENSRNOT00000081141 |
Dnajc27
|
DnaJ heat shock protein family (Hsp40) member C27 |
| chr17_+_10463303 | 0.07 |
ENSRNOT00000060822
|
Rnf44
|
ring finger protein 44 |
| chr6_-_41870046 | 0.07 |
ENSRNOT00000005863
|
Lpin1
|
lipin 1 |
| chr4_+_157326727 | 0.07 |
ENSRNOT00000020493
|
Spsb2
|
splA/ryanodine receptor domain and SOCS box containing 2 |
| chr4_-_88565292 | 0.07 |
ENSRNOT00000008948
|
Lancl2
|
LanC like 2 |
| chr3_-_9236736 | 0.06 |
ENSRNOT00000072628
|
Nup214
|
nucleoporin 214 |
| chr9_-_20219209 | 0.06 |
ENSRNOT00000072086
|
LOC100911548
|
SPRY domain-containing SOCS box protein 2-like |
| chr9_+_19451630 | 0.05 |
ENSRNOT00000065048
|
Enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr16_-_69195097 | 0.05 |
ENSRNOT00000018973
|
Erlin2
|
ER lipid raft associated 2 |
| chr5_-_152464850 | 0.05 |
ENSRNOT00000021937
|
Zfp593
|
zinc finger protein 593 |
| chr11_-_64752544 | 0.05 |
ENSRNOT00000048738
|
Tmem39a
|
transmembrane protein 39a |
| chr1_-_156296161 | 0.05 |
ENSRNOT00000025653
|
Crebzf
|
CREB/ATF bZIP transcription factor |
| chr8_+_114867062 | 0.05 |
ENSRNOT00000074771
|
Wdr82
|
WD repeat domain 82 |
| chr7_-_18612118 | 0.04 |
ENSRNOT00000078122
ENSRNOT00000010197 |
Rab11b
|
RAB11B, member RAS oncogene family |
| chr3_+_112371677 | 0.04 |
ENSRNOT00000013316
|
LOC100912076
|
HAUS augmin-like complex subunit 2-like |
| chr4_-_6330699 | 0.04 |
ENSRNOT00000078077
ENSRNOT00000011814 |
Galnt11
|
polypeptide N-acetylgalactosaminyltransferase 11 |
| chr20_-_10257044 | 0.04 |
ENSRNOT00000068289
|
Wdr4
|
WD repeat domain 4 |
| chr15_-_34392066 | 0.04 |
ENSRNOT00000027315
|
Tgm1
|
transglutaminase 1 |
| chr6_+_96171756 | 0.04 |
ENSRNOT00000029328
|
Slc38a6
|
solute carrier family 38, member 6 |
| chr2_+_189629297 | 0.04 |
ENSRNOT00000049810
|
Dennd4b
|
DENN domain containing 4B |
| chr19_+_56272162 | 0.04 |
ENSRNOT00000030399
|
Afg3l1
|
AFG3(ATPase family gene 3)-like 1 (S. cerevisiae) |
| chr12_-_2170504 | 0.03 |
ENSRNOT00000001309
|
Xab2
|
XPA binding protein 2 |
| chr5_+_148577332 | 0.03 |
ENSRNOT00000016325
|
Snrnp40
|
small nuclear ribonucleoprotein U5 subunit 40 |
| chr2_+_240396152 | 0.03 |
ENSRNOT00000034565
|
Cenpe
|
centromere protein E |
| chr4_-_157078130 | 0.03 |
ENSRNOT00000015570
|
Clstn3
|
calsyntenin 3 |
| chrX_-_158978995 | 0.02 |
ENSRNOT00000001179
|
Mmgt1
|
membrane magnesium transporter 1 |
| chr3_-_117389456 | 0.02 |
ENSRNOT00000007103
ENSRNOT00000081533 |
Myef2
|
myelin expression factor 2 |
| chr17_+_10537365 | 0.02 |
ENSRNOT00000023651
|
Cltb
|
clathrin, light chain B |
| chr3_+_148635775 | 0.02 |
ENSRNOT00000067261
|
Tm9sf4
|
transmembrane 9 superfamily member 4 |
| chr10_+_11724032 | 0.02 |
ENSRNOT00000008966
|
Trap1
|
TNF receptor-associated protein 1 |
| chr1_+_201256910 | 0.02 |
ENSRNOT00000090143
|
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr1_+_78015722 | 0.01 |
ENSRNOT00000002043
|
Kptn
|
kaptin (actin binding protein) |
| chr2_-_185005572 | 0.01 |
ENSRNOT00000093291
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr12_+_13102019 | 0.01 |
ENSRNOT00000092628
|
Rac1
|
ras-related C3 botulinum toxin substrate 1 |
| chr2_-_104461863 | 0.01 |
ENSRNOT00000016953
|
Crh
|
corticotropin releasing hormone |
| chr1_-_259691742 | 0.01 |
ENSRNOT00000088065
|
Tctn3
|
tectonic family member 3 |
| chr2_-_66608324 | 0.01 |
ENSRNOT00000077597
|
Cln5
|
ceroid-lipofuscinosis, neuronal 5 |
| chr1_-_259106948 | 0.01 |
ENSRNOT00000074429
|
LOC100912537
|
tectonic-3-like |
| chr3_-_120011364 | 0.01 |
ENSRNOT00000018922
|
Fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr12_+_17416327 | 0.01 |
ENSRNOT00000089590
ENSRNOT00000092186 |
Adap1
|
ArfGAP with dual PH domains 1 |
| chr5_+_159845774 | 0.00 |
ENSRNOT00000012328
|
Epha2
|
Eph receptor A2 |
| chr1_+_261415191 | 0.00 |
ENSRNOT00000083287
ENSRNOT00000040740 |
Zfyve27
|
zinc finger FYVE-type containing 27 |
| chr4_+_113887115 | 0.00 |
ENSRNOT00000010602
|
Aup1
|
ancient ubiquitous protein 1 |
| chr14_+_1462358 | 0.00 |
ENSRNOT00000077243
|
Csf2ra
|
colony stimulating factor 2 receptor alpha subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of Batf3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.0 | 0.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.4 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.0 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.0 | GO:0005745 | m-AAA complex(GO:0005745) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0031177 | phosphopantetheine binding(GO:0031177) |