Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
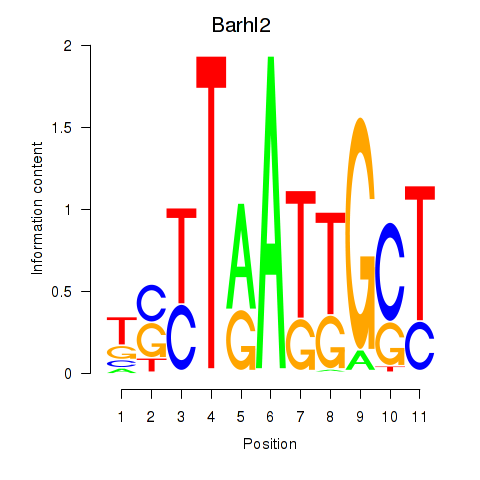
Results for Barhl2
Z-value: 0.20
Transcription factors associated with Barhl2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Barhl2
|
ENSRNOG00000002117 | BarH-like homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Barhl2 | rn6_v1_chr14_+_4362717_4362717 | -0.61 | 2.8e-01 | Click! |
Activity profile of Barhl2 motif
Sorted Z-values of Barhl2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_260596777 | 0.08 |
ENSRNOT00000009370
|
Lhx8
|
LIM homeobox 8 |
| chr3_-_122206671 | 0.07 |
ENSRNOT00000037576
|
Pdyn
|
prodynorphin |
| chr17_-_43807540 | 0.05 |
ENSRNOT00000074763
|
LOC684762
|
similar to CG31613-PA |
| chr8_+_49441106 | 0.05 |
ENSRNOT00000030152
|
Scn4b
|
sodium voltage-gated channel beta subunit 4 |
| chr14_-_114047527 | 0.05 |
ENSRNOT00000083199
|
AABR07016779.1
|
|
| chr15_+_48674380 | 0.05 |
ENSRNOT00000018762
|
Fbxo16
|
F-box protein 16 |
| chr3_-_10196626 | 0.05 |
ENSRNOT00000012234
|
Prdm12
|
PR/SET domain 12 |
| chr12_-_19254527 | 0.04 |
ENSRNOT00000089349
|
AABR07035541.3
|
|
| chr16_-_49453394 | 0.04 |
ENSRNOT00000041617
|
Lrp2bp
|
Lrp2 binding protein |
| chr11_-_83483513 | 0.04 |
ENSRNOT00000084380
|
AABR07034673.1
|
|
| chr6_+_3375476 | 0.04 |
ENSRNOT00000045349
|
LOC102551744
|
eukaryotic translation initiation factor 1-like |
| chr5_-_136098013 | 0.04 |
ENSRNOT00000089166
|
RGD1563714
|
RGD1563714 |
| chr3_+_147422095 | 0.04 |
ENSRNOT00000006786
|
Angpt4
|
angiopoietin 4 |
| chr2_+_210880777 | 0.04 |
ENSRNOT00000026237
|
Gnat2
|
G protein subunit alpha transducin 2 |
| chr17_-_78735324 | 0.03 |
ENSRNOT00000036299
|
Cdnf
|
cerebral dopamine neurotrophic factor |
| chr7_+_143122269 | 0.03 |
ENSRNOT00000082542
ENSRNOT00000045495 ENSRNOT00000081386 ENSRNOT00000067422 |
Krt86
|
keratin 86 |
| chr18_+_57727187 | 0.03 |
ENSRNOT00000065483
ENSRNOT00000025871 |
Htr4
|
5-hydroxytryptamine receptor 4 |
| chr10_-_25910298 | 0.03 |
ENSRNOT00000065633
ENSRNOT00000079646 |
Ccng1
|
cyclin G1 |
| chr13_-_111972603 | 0.03 |
ENSRNOT00000007870
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr9_+_40817654 | 0.03 |
ENSRNOT00000037392
|
AABR07067339.1
|
|
| chr7_-_143228060 | 0.03 |
ENSRNOT00000088923
ENSRNOT00000012640 |
Krt75
|
keratin 75 |
| chr10_-_48038647 | 0.03 |
ENSRNOT00000078448
|
Prpsap2
|
phosphoribosyl pyrophosphate synthetase-associated protein 2 |
| chr4_-_178168690 | 0.03 |
ENSRNOT00000020729
|
Sox5
|
SRY box 5 |
| chr20_-_4070721 | 0.03 |
ENSRNOT00000000523
|
RT1-Ba
|
RT1 class II, locus Ba |
| chr1_-_229639187 | 0.03 |
ENSRNOT00000016804
|
Zfp91
|
zinc finger protein 91 |
| chr7_-_145338152 | 0.03 |
ENSRNOT00000055268
|
Spt1
|
salivary protein 1 |
| chr5_-_159946446 | 0.03 |
ENSRNOT00000089184
|
Clcnka
|
chloride voltage-gated channel Ka |
| chrX_-_25628272 | 0.03 |
ENSRNOT00000086414
|
Mid1
|
midline 1 |
| chr7_+_141249044 | 0.03 |
ENSRNOT00000084911
|
Aqp5
|
aquaporin 5 |
| chr11_-_64952687 | 0.03 |
ENSRNOT00000087892
|
Popdc2
|
popeye domain containing 2 |
| chr7_-_145575981 | 0.02 |
ENSRNOT00000074622
|
LOC102555445
|
16.5 kDa submandibular gland glycoprotein-like |
| chr15_+_4850122 | 0.02 |
ENSRNOT00000071133
|
Gng2
|
G protein subunit gamma 2 |
| chr12_-_51877624 | 0.02 |
ENSRNOT00000056800
|
Chek2
|
checkpoint kinase 2 |
| chr2_+_189609800 | 0.02 |
ENSRNOT00000089016
|
Slc39a1
|
solute carrier family 39 member 1 |
| chr9_-_55673704 | 0.02 |
ENSRNOT00000066231
ENSRNOT00000081677 |
Tmeff2
|
transmembrane protein with EGF-like and two follistatin-like domains 2 |
| chr18_+_81694808 | 0.02 |
ENSRNOT00000020446
|
Cyb5a
|
cytochrome b5 type A |
| chr5_+_150957001 | 0.02 |
ENSRNOT00000017549
|
Rpa2
|
replication protein A2 |
| chr5_+_144106802 | 0.02 |
ENSRNOT00000035637
ENSRNOT00000079779 |
Lsm10
|
LSM10, U7 small nuclear RNA associated |
| chrX_+_71335491 | 0.02 |
ENSRNOT00000076003
|
Nono
|
non-POU domain containing, octamer-binding |
| chr2_-_179704629 | 0.02 |
ENSRNOT00000083361
ENSRNOT00000077941 |
Gria2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr15_+_44441856 | 0.02 |
ENSRNOT00000018006
|
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr7_-_49741540 | 0.02 |
ENSRNOT00000006523
|
Myf6
|
myogenic factor 6 |
| chr2_+_197720259 | 0.02 |
ENSRNOT00000070919
|
Golph3l
|
golgi phosphoprotein 3-like |
| chr2_+_23289374 | 0.02 |
ENSRNOT00000090666
ENSRNOT00000032783 |
Dmgdh
|
dimethylglycine dehydrogenase |
| chr15_-_83494423 | 0.02 |
ENSRNOT00000037588
|
Dis3
|
DIS3 homolog, exosome endoribonuclease and 3'-5' exoribonuclease |
| chr9_-_74048244 | 0.02 |
ENSRNOT00000018293
|
Lancl1
|
LanC like 1 |
| chr3_-_46601409 | 0.02 |
ENSRNOT00000079261
|
Pla2r1
|
phospholipase A2 receptor 1 |
| chr10_-_87248572 | 0.02 |
ENSRNOT00000066637
ENSRNOT00000085677 |
Krt26
|
keratin 26 |
| chr5_-_115387377 | 0.02 |
ENSRNOT00000036030
ENSRNOT00000077492 |
RGD1560146
|
similar to hypothetical protein MGC34837 |
| chr1_-_64021321 | 0.02 |
ENSRNOT00000090819
|
Rps9
|
ribosomal protein S9 |
| chrX_-_115496045 | 0.02 |
ENSRNOT00000051134
|
LOC100361854
|
ribosomal protein S26-like |
| chr9_+_81644355 | 0.02 |
ENSRNOT00000071700
|
Catip
|
ciliogenesis associated TTC17 interacting protein |
| chr10_-_86932154 | 0.01 |
ENSRNOT00000085344
|
Top2a
|
topoisomerase (DNA) II alpha |
| chr1_+_82169620 | 0.01 |
ENSRNOT00000088955
ENSRNOT00000068251 |
Prr19
|
proline rich 19 |
| chr2_-_112831476 | 0.01 |
ENSRNOT00000018055
|
Ect2
|
epithelial cell transforming 2 |
| chr4_+_2055615 | 0.01 |
ENSRNOT00000007646
|
Rnf32
|
ring finger protein 32 |
| chr17_+_17965881 | 0.01 |
ENSRNOT00000021751
|
Dek
|
DEK proto-oncogene |
| chr10_+_34402482 | 0.01 |
ENSRNOT00000072317
|
Tpcr12
|
putative olfactory receptor |
| chr9_+_3238077 | 0.01 |
ENSRNOT00000088083
|
RGD1559808
|
similar to 40S ribosomal protein S26 |
| chr16_+_14306804 | 0.01 |
ENSRNOT00000017773
|
Lrit1
|
leucine-rich repeat, Ig-like and transmembrane domains 1 |
| chr1_+_64162672 | 0.01 |
ENSRNOT00000089713
|
Tfpt
|
TCF3 (E2A) fusion partner |
| chr20_-_157665 | 0.01 |
ENSRNOT00000048858
ENSRNOT00000079494 |
RT1-CE10
|
RT1 class I, locus CE10 |
| chr13_-_68360664 | 0.01 |
ENSRNOT00000030971
|
Hmcn1
|
hemicentin 1 |
| chr10_+_43851950 | 0.01 |
ENSRNOT00000085666
|
LOC691277
|
similar to Robo-1 |
| chr13_+_47572219 | 0.01 |
ENSRNOT00000088449
ENSRNOT00000087664 ENSRNOT00000005853 |
Pigr
|
polymeric immunoglobulin receptor |
| chr20_+_44680449 | 0.01 |
ENSRNOT00000000728
|
Traf3ip2
|
Traf3 interacting protein 2 |
| chr13_-_84452181 | 0.01 |
ENSRNOT00000005060
|
Mael
|
maelstrom spermatogenic transposon silencer |
| chr10_-_15098791 | 0.01 |
ENSRNOT00000026139
|
Chtf18
|
chromosome transmission fidelity factor 18 |
| chr11_-_32550539 | 0.01 |
ENSRNOT00000002715
|
Rcan1
|
regulator of calcineurin 1 |
| chr6_+_69971227 | 0.01 |
ENSRNOT00000075349
|
Foxg1
|
forkhead box G1 |
| chr1_+_81230612 | 0.01 |
ENSRNOT00000026489
|
Kcnn4
|
potassium calcium-activated channel subfamily N member 4 |
| chr1_-_238381871 | 0.01 |
ENSRNOT00000077601
|
Tmc1
|
transmembrane channel-like 1 |
| chr10_-_109747987 | 0.01 |
ENSRNOT00000054958
|
P4hb
|
prolyl 4-hydroxylase subunit beta |
| chr20_-_157861 | 0.01 |
ENSRNOT00000084461
|
RT1-CE10
|
RT1 class I, locus CE10 |
| chr8_+_60117729 | 0.01 |
ENSRNOT00000021074
|
Isl2
|
ISL LIM homeobox 2 |
| chr11_+_73930753 | 0.01 |
ENSRNOT00000087747
|
RGD1562415
|
similar to 40S ribosomal protein S26 |
| chr17_+_78735598 | 0.01 |
ENSRNOT00000020854
|
Hspa14
|
heat shock protein family A, member 14 |
| chr5_+_57845819 | 0.01 |
ENSRNOT00000017712
|
Nudt2
|
nudix hydrolase 2 |
| chr5_-_58445953 | 0.01 |
ENSRNOT00000046102
|
Vcp
|
valosin-containing protein |
| chr3_+_100768637 | 0.01 |
ENSRNOT00000083542
|
Bdnf
|
brain-derived neurotrophic factor |
| chr1_-_185143272 | 0.01 |
ENSRNOT00000027752
|
Nucb2
|
nucleobindin 2 |
| chr1_+_199287895 | 0.01 |
ENSRNOT00000026224
|
Stx4
|
syntaxin 4 |
| chr3_-_165700489 | 0.01 |
ENSRNOT00000017008
|
Zfp93
|
zinc finger protein 93 |
| chr2_+_18392142 | 0.01 |
ENSRNOT00000043196
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr13_-_83425641 | 0.01 |
ENSRNOT00000063870
|
Tbx19
|
T-box 19 |
| chr1_-_247821728 | 0.01 |
ENSRNOT00000021958
|
Ermp1
|
endoplasmic reticulum metallopeptidase 1 |
| chr15_-_49467174 | 0.01 |
ENSRNOT00000019209
|
Adam7
|
ADAM metallopeptidase domain 7 |
| chr2_+_154580558 | 0.01 |
ENSRNOT00000076976
|
Gmps
|
guanine monophosphate synthase |
| chr8_-_12355091 | 0.01 |
ENSRNOT00000009318
|
Cep57
|
centrosomal protein 57 |
| chr18_+_17091310 | 0.01 |
ENSRNOT00000093285
|
Fhod3
|
formin homology 2 domain containing 3 |
| chr1_+_84293102 | 0.00 |
ENSRNOT00000028468
|
Sertad1
|
SERTA domain containing 1 |
| chr20_+_4369394 | 0.00 |
ENSRNOT00000088734
ENSRNOT00000091491 |
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr3_+_117421604 | 0.00 |
ENSRNOT00000008860
ENSRNOT00000008857 |
Slc12a1
|
solute carrier family 12 member 1 |
| chr14_-_21299068 | 0.00 |
ENSRNOT00000065778
|
Amtn
|
amelotin |
| chr5_-_159962218 | 0.00 |
ENSRNOT00000050729
|
Clcnkb
|
chloride voltage-gated channel Kb |
| chr1_-_164101578 | 0.00 |
ENSRNOT00000022176
|
Uvrag
|
UV radiation resistance associated |
| chr1_+_84328114 | 0.00 |
ENSRNOT00000028266
|
Hipk4
|
homeodomain interacting protein kinase 4 |
| chr3_-_44086006 | 0.00 |
ENSRNOT00000034449
ENSRNOT00000082604 |
Ermn
|
ermin |
| chr11_-_782954 | 0.00 |
ENSRNOT00000040065
|
Epha3
|
Eph receptor A3 |
| chr1_-_64090017 | 0.00 |
ENSRNOT00000086622
ENSRNOT00000091654 |
Rps9
|
ribosomal protein S9 |
| chr3_-_125470413 | 0.00 |
ENSRNOT00000028893
|
Trmt6
|
tRNA methyltransferase 6 |
| chr7_-_117440342 | 0.00 |
ENSRNOT00000018528
|
Tssk5
|
testis-specific serine kinase 5 |
| chr20_-_5533600 | 0.00 |
ENSRNOT00000072319
|
Cuta
|
cutA divalent cation tolerance homolog |
| chr14_+_43512901 | 0.00 |
ENSRNOT00000050664
|
Apbb2
|
amyloid beta precursor protein binding family B member 2 |
| chr3_-_171342646 | 0.00 |
ENSRNOT00000071853
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr20_-_5533448 | 0.00 |
ENSRNOT00000000568
|
Cuta
|
cutA divalent cation tolerance homolog |
| chr6_-_92121351 | 0.00 |
ENSRNOT00000006374
|
Cdkl1
|
cyclin dependent kinase like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Barhl2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.0 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |