Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
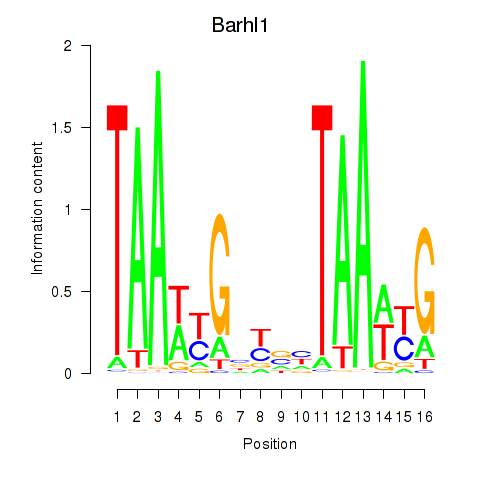
Results for Barhl1
Z-value: 0.26
Transcription factors associated with Barhl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Barhl1
|
ENSRNOG00000013209 | BarH-like homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Barhl1 | rn6_v1_chr3_-_7498555_7498555 | -0.85 | 7.1e-02 | Click! |
Activity profile of Barhl1 motif
Sorted Z-values of Barhl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_53220397 | 0.12 |
ENSRNOT00000089989
|
AABR07001573.1
|
|
| chr8_-_104155775 | 0.11 |
ENSRNOT00000042885
|
LOC102550734
|
60S ribosomal protein L31-like |
| chr13_+_91054974 | 0.11 |
ENSRNOT00000091089
|
Crp
|
C-reactive protein |
| chr7_+_143122269 | 0.10 |
ENSRNOT00000082542
ENSRNOT00000045495 ENSRNOT00000081386 ENSRNOT00000067422 |
Krt86
|
keratin 86 |
| chr16_+_22979444 | 0.08 |
ENSRNOT00000017822
|
Csgalnact1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr2_-_30340103 | 0.05 |
ENSRNOT00000024023
ENSRNOT00000067722 |
Bdp1
|
B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB |
| chr4_-_66624912 | 0.05 |
ENSRNOT00000064891
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr9_+_73334618 | 0.05 |
ENSRNOT00000092717
|
Map2
|
microtubule-associated protein 2 |
| chr4_+_22859622 | 0.05 |
ENSRNOT00000073501
ENSRNOT00000068410 |
Adam22
|
ADAM metallopeptidase domain 22 |
| chr3_+_154437571 | 0.04 |
ENSRNOT00000019331
|
AABR07054456.1
|
|
| chr9_-_63641400 | 0.04 |
ENSRNOT00000087684
|
Satb2
|
SATB homeobox 2 |
| chrX_+_14578264 | 0.04 |
ENSRNOT00000038994
|
Cybb
|
cytochrome b-245 beta chain |
| chr17_-_52477575 | 0.04 |
ENSRNOT00000081290
|
Gli3
|
GLI family zinc finger 3 |
| chr4_-_85174931 | 0.04 |
ENSRNOT00000014324
ENSRNOT00000088085 |
Nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chr1_+_205911552 | 0.04 |
ENSRNOT00000024751
|
Fank1
|
fibronectin type 3 and ankyrin repeat domains 1 |
| chr2_-_243475639 | 0.03 |
ENSRNOT00000089222
|
RGD1309170
|
similar to hypothetical protein DKFZp434G072 |
| chr6_-_50941248 | 0.03 |
ENSRNOT00000084533
|
Dus4l
|
dihydrouridine synthase 4-like |
| chrX_+_123751089 | 0.03 |
ENSRNOT00000092384
|
Nkap
|
NFKB activating protein |
| chr11_-_88888377 | 0.03 |
ENSRNOT00000034902
|
Yars2
|
tyrosyl-tRNA synthetase 2 |
| chr8_+_5606592 | 0.03 |
ENSRNOT00000011727
|
Mmp12
|
matrix metallopeptidase 12 |
| chr14_+_17210733 | 0.03 |
ENSRNOT00000003075
|
Cxcl10
|
C-X-C motif chemokine ligand 10 |
| chr2_+_69415057 | 0.03 |
ENSRNOT00000013152
|
Cdh10
|
cadherin 10 |
| chr1_+_266844480 | 0.03 |
ENSRNOT00000027482
|
Taf5
|
TATA-box binding protein associated factor 5 |
| chr8_+_128972311 | 0.03 |
ENSRNOT00000025460
|
Myrip
|
myosin VIIA and Rab interacting protein |
| chr20_-_32296840 | 0.03 |
ENSRNOT00000080727
|
Stox1
|
storkhead box 1 |
| chr3_+_104749051 | 0.02 |
ENSRNOT00000000010
|
Tmco5b
|
transmembrane and coiled-coil domains 5B |
| chr11_-_45124423 | 0.02 |
ENSRNOT00000046383
|
Filip1l
|
filamin A interacting protein 1-like |
| chr14_+_59611434 | 0.02 |
ENSRNOT00000065366
|
Cckar
|
cholecystokinin A receptor |
| chr3_+_103753238 | 0.02 |
ENSRNOT00000007144
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr16_-_3765917 | 0.02 |
ENSRNOT00000088284
|
Duxbl1
|
double homeobox B-like 1 |
| chr1_-_140835151 | 0.02 |
ENSRNOT00000032174
|
Hapln3
|
hyaluronan and proteoglycan link protein 3 |
| chr18_+_4325875 | 0.02 |
ENSRNOT00000073771
|
Impact
|
impact RWD domain protein |
| chr16_-_36161089 | 0.02 |
ENSRNOT00000017888
|
Scrg1
|
stimulator of chondrogenesis 1 |
| chr3_-_104502471 | 0.02 |
ENSRNOT00000040306
|
Ryr3
|
ryanodine receptor 3 |
| chr3_-_8615532 | 0.02 |
ENSRNOT00000021132
|
Wdr34
|
WD repeat domain 34 |
| chr6_-_75670135 | 0.02 |
ENSRNOT00000007300
|
Snx6
|
sorting nexin 6 |
| chr3_+_114102875 | 0.02 |
ENSRNOT00000023209
|
Trim69
|
tripartite motif-containing 69 |
| chr3_+_114900343 | 0.02 |
ENSRNOT00000068129
|
Sqrdl
|
sulfide quinone reductase-like (yeast) |
| chr2_+_198721724 | 0.02 |
ENSRNOT00000043535
|
Ankrd34a
|
ankyrin repeat domain 34A |
| chr10_-_65766050 | 0.01 |
ENSRNOT00000013639
|
Sarm1
|
sterile alpha and TIR motif containing 1 |
| chr13_+_42220251 | 0.01 |
ENSRNOT00000078185
|
AABR07020815.1
|
|
| chr10_+_10530365 | 0.01 |
ENSRNOT00000003843
|
Eef2kmt
|
eukaryotic elongation factor 2 lysine methyltransferase |
| chr8_+_85503224 | 0.01 |
ENSRNOT00000012348
|
Gsta4
|
glutathione S-transferase alpha 4 |
| chr10_+_90377103 | 0.01 |
ENSRNOT00000040472
|
Grn
|
granulin precursor |
| chr10_+_90376933 | 0.01 |
ENSRNOT00000028557
|
Grn
|
granulin precursor |
| chr4_-_70934295 | 0.01 |
ENSRNOT00000020616
|
Trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr4_-_183374195 | 0.01 |
ENSRNOT00000073927
ENSRNOT00000074740 ENSRNOT00000070907 |
Caprin2
|
caprin family member 2 |
| chr2_-_170301348 | 0.01 |
ENSRNOT00000088131
|
Si
|
sucrase-isomaltase |
| chr18_+_29993361 | 0.01 |
ENSRNOT00000075810
|
Pcdha4
|
protocadherin alpha 4 |
| chr12_+_12859661 | 0.01 |
ENSRNOT00000001404
|
Usp42
|
ubiquitin specific peptidase 42 |
| chr3_-_59166356 | 0.01 |
ENSRNOT00000047713
|
AABR07052519.1
|
|
| chrX_-_118459355 | 0.01 |
ENSRNOT00000084465
|
Il13ra2
|
interleukin 13 receptor subunit alpha 2 |
| chr1_+_185863043 | 0.01 |
ENSRNOT00000079072
|
Sox6
|
SRY box 6 |
| chr10_-_55851235 | 0.01 |
ENSRNOT00000010790
|
Gucy2d
|
guanylate cyclase 2D, retinal |
| chr20_+_3992496 | 0.01 |
ENSRNOT00000082047
|
Psmb8
|
proteasome subunit beta 8 |
| chr3_+_101791337 | 0.01 |
ENSRNOT00000046937
|
Slc5a12
|
solute carrier family 5 member 12 |
| chr3_-_112985318 | 0.01 |
ENSRNOT00000015556
|
Epb42
|
erythrocyte membrane protein band 4.2 |
| chr13_+_47572219 | 0.01 |
ENSRNOT00000088449
ENSRNOT00000087664 ENSRNOT00000005853 |
Pigr
|
polymeric immunoglobulin receptor |
| chr4_+_31333970 | 0.01 |
ENSRNOT00000064866
|
LOC100911994
|
coiled-coil domain-containing protein 132-like |
| chr1_-_73732118 | 0.01 |
ENSRNOT00000077964
|
Leng8
|
leukocyte receptor cluster member 8 |
| chr15_+_106434177 | 0.01 |
ENSRNOT00000015271
|
Farp1
|
FERM, ARH/RhoGEF and pleckstrin domain protein 1 |
| chr17_+_47302272 | 0.01 |
ENSRNOT00000090021
|
Nme8
|
NME/NM23 family member 8 |
| chr5_+_61657507 | 0.01 |
ENSRNOT00000013228
|
Tmod1
|
tropomodulin 1 |
| chr11_+_43194348 | 0.01 |
ENSRNOT00000075303
|
Olr1532
|
olfactory receptor 1532 |
| chr19_-_25955371 | 0.01 |
ENSRNOT00000004042
ENSRNOT00000084123 |
Rad23a
|
RAD23 homolog A, nucleotide excision repair protein |
| chr20_-_46044738 | 0.00 |
ENSRNOT00000000343
|
Fig4
|
FIG4 phosphoinositide 5-phosphatase |
| chr8_-_55171718 | 0.00 |
ENSRNOT00000080736
|
LOC689959
|
hypothetical protein LOC689959 |
| chr1_-_73226777 | 0.00 |
ENSRNOT00000025080
|
Ncr1
|
natural cytotoxicity triggering receptor 1 |
| chr9_+_47536824 | 0.00 |
ENSRNOT00000049349
|
Tmem182
|
transmembrane protein 182 |
| chr4_-_164406146 | 0.00 |
ENSRNOT00000090110
|
Klra22
|
killer cell lectin-like receptor subfamily A, member 22 |
| chr10_+_96661696 | 0.00 |
ENSRNOT00000077567
|
AABR07030630.1
|
|
| chr9_-_82477136 | 0.00 |
ENSRNOT00000026753
ENSRNOT00000089555 |
Resp18
|
regulated endocrine-specific protein 18 |
| chr3_-_61488696 | 0.00 |
ENSRNOT00000083045
|
Lnpk
|
lunapark, ER junction formation factor |
| chr1_-_165606375 | 0.00 |
ENSRNOT00000024290
|
Mrpl48
|
mitochondrial ribosomal protein L48 |
| chr10_-_90127600 | 0.00 |
ENSRNOT00000028368
|
Lsm12
|
LSM12 homolog |
| chr18_-_36579403 | 0.00 |
ENSRNOT00000025284
|
Lars
|
leucyl-tRNA synthetase |
| chr1_+_279896973 | 0.00 |
ENSRNOT00000068119
|
Pnliprp2
|
pancreatic lipase related protein 2 |
| chr3_+_94549180 | 0.00 |
ENSRNOT00000016318
|
Cstf3
|
cleavage stimulation factor subunit 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Barhl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.0 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) TFIIIC-class transcription factor binding(GO:0001156) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |