Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
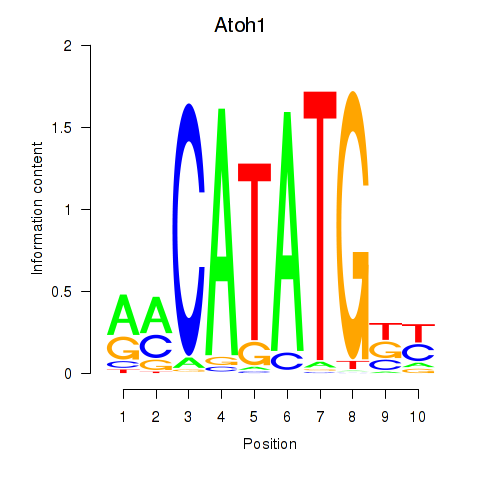
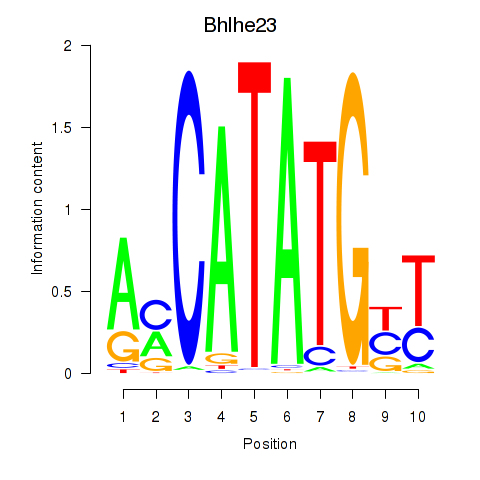
Results for Atoh1_Bhlhe23
Z-value: 0.56
Transcription factors associated with Atoh1_Bhlhe23
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Atoh1
|
ENSRNOG00000006383 | atonal bHLH transcription factor 1 |
|
Bhlhe23
|
ENSRNOG00000010312 | basic helix-loop-helix family, member e23 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atoh1 | rn6_v1_chr4_+_95498003_95498003 | 0.88 | 5.1e-02 | Click! |
| Bhlhe23 | rn6_v1_chr3_-_176282211_176282211 | 0.69 | 1.9e-01 | Click! |
Activity profile of Atoh1_Bhlhe23 motif
Sorted Z-values of Atoh1_Bhlhe23 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_66417741 | 0.75 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr10_-_63952726 | 0.33 |
ENSRNOT00000090461
|
Doc2b
|
double C2 domain beta |
| chr8_+_69971778 | 0.26 |
ENSRNOT00000058007
ENSRNOT00000037941 ENSRNOT00000050649 |
Megf11
|
multiple EGF-like-domains 11 |
| chr7_-_114848414 | 0.26 |
ENSRNOT00000032620
|
Slc45a4
|
solute carrier family 45, member 4 |
| chr5_-_12563429 | 0.23 |
ENSRNOT00000059625
|
St18
|
suppression of tumorigenicity 18 |
| chr8_-_63750531 | 0.20 |
ENSRNOT00000009496
|
Neo1
|
neogenin 1 |
| chr4_-_31730386 | 0.20 |
ENSRNOT00000013817
|
Slc25a13
|
solute carrier family 25 member 13 |
| chr7_-_117978787 | 0.19 |
ENSRNOT00000074262
|
Zfp647
|
zinc finger protein 647 |
| chr3_-_66335869 | 0.17 |
ENSRNOT00000079781
ENSRNOT00000043238 ENSRNOT00000073412 ENSRNOT00000057878 ENSRNOT00000079212 |
Cerkl
|
ceramide kinase-like |
| chr5_-_12526962 | 0.17 |
ENSRNOT00000092104
|
St18
|
suppression of tumorigenicity 18 |
| chr2_-_177924970 | 0.17 |
ENSRNOT00000029340
|
Rapgef2
|
Rap guanine nucleotide exchange factor 2 |
| chr10_+_74959285 | 0.16 |
ENSRNOT00000010296
|
Rnf43
|
ring finger protein 43 |
| chr10_-_45297385 | 0.16 |
ENSRNOT00000041187
|
Hist3h2bb
|
histone cluster 3 H2B family member b |
| chr2_+_196334626 | 0.15 |
ENSRNOT00000050914
ENSRNOT00000028645 ENSRNOT00000090729 |
Sema6c
|
semaphorin 6C |
| chr6_+_60566196 | 0.15 |
ENSRNOT00000006709
ENSRNOT00000075193 |
Dock4
|
dedicator of cytokinesis 4 |
| chr10_-_56530842 | 0.15 |
ENSRNOT00000077451
|
AABR07029863.3
|
|
| chr4_-_81241152 | 0.13 |
ENSRNOT00000015325
ENSRNOT00000015152 |
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr6_+_52663112 | 0.13 |
ENSRNOT00000013842
|
Atxn7l1
|
ataxin 7-like 1 |
| chr3_+_145764932 | 0.12 |
ENSRNOT00000075257
|
AABR07054264.1
|
|
| chr3_+_40038336 | 0.12 |
ENSRNOT00000048804
|
Galnt13
|
polypeptide N-acetylgalactosaminyltransferase 13 |
| chr2_+_207930796 | 0.11 |
ENSRNOT00000047827
|
Kcnd3
|
potassium voltage-gated channel subfamily D member 3 |
| chr4_-_130659697 | 0.11 |
ENSRNOT00000072374
|
LOC100363436
|
rCG56280-like |
| chr10_-_47724499 | 0.11 |
ENSRNOT00000085011
|
Rnf112
|
ring finger protein 112 |
| chrX_-_76708878 | 0.10 |
ENSRNOT00000045534
|
Atrx
|
ATRX, chromatin remodeler |
| chr17_+_66548818 | 0.10 |
ENSRNOT00000024361
|
Anlnl1
|
anillin, actin binding protein-like 1 |
| chr13_-_88642011 | 0.09 |
ENSRNOT00000067037
|
LOC100361087
|
hypothetical LOC100361087 |
| chr3_+_172155496 | 0.09 |
ENSRNOT00000066279
|
Stx16
|
syntaxin 16 |
| chr9_+_16302578 | 0.09 |
ENSRNOT00000021762
|
Gltscr1l
|
GLTSCR1-like |
| chr10_-_16046033 | 0.08 |
ENSRNOT00000089460
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr19_+_20147037 | 0.08 |
ENSRNOT00000020028
|
Zfp423
|
zinc finger protein 423 |
| chr10_-_16045835 | 0.08 |
ENSRNOT00000064832
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr9_-_81586116 | 0.08 |
ENSRNOT00000089952
|
Tmbim1
|
transmembrane BAX inhibitor motif containing 1 |
| chr9_+_51298426 | 0.08 |
ENSRNOT00000065212
|
Gulp1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr5_+_98469047 | 0.07 |
ENSRNOT00000041374
|
Lurap1l
|
leucine rich adaptor protein 1-like |
| chr11_+_9642365 | 0.07 |
ENSRNOT00000087080
ENSRNOT00000042384 |
Robo1
|
roundabout guidance receptor 1 |
| chr3_+_128756799 | 0.07 |
ENSRNOT00000049855
ENSRNOT00000042853 |
Plcb4
|
phospholipase C, beta 4 |
| chrX_-_76924362 | 0.07 |
ENSRNOT00000078997
|
Atrx
|
ATRX, chromatin remodeler |
| chr7_-_118518193 | 0.07 |
ENSRNOT00000075636
|
LOC102555083
|
zinc finger protein 250-like |
| chr6_-_10515370 | 0.07 |
ENSRNOT00000020845
|
Atp6v1e2
|
ATPase H+ transporting V1 subunit E2 |
| chr20_+_6211420 | 0.07 |
ENSRNOT00000000624
|
Kctd20
|
potassium channel tetramerization domain containing 20 |
| chr3_+_45683993 | 0.06 |
ENSRNOT00000038983
|
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr1_-_198577226 | 0.06 |
ENSRNOT00000055013
|
Spn
|
sialophorin |
| chr20_-_5212624 | 0.06 |
ENSRNOT00000074261
|
LOC103689996
|
antigen peptide transporter 2 |
| chr2_+_123617794 | 0.06 |
ENSRNOT00000080632
|
RGD1307100
|
similar to RIKEN cDNA D630029K19 |
| chr2_+_196594303 | 0.06 |
ENSRNOT00000064442
ENSRNOT00000044738 |
Arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr10_+_84182118 | 0.06 |
ENSRNOT00000011027
|
Hoxb3
|
homeo box B3 |
| chr2_-_118989127 | 0.06 |
ENSRNOT00000014871
|
Gnb4
|
G protein subunit beta 4 |
| chr5_+_58636083 | 0.06 |
ENSRNOT00000067281
|
Unc13b
|
unc-13 homolog B |
| chr2_+_122368265 | 0.06 |
ENSRNOT00000078321
|
Atp11b
|
ATPase phospholipid transporting 11B (putative) |
| chr6_-_76552559 | 0.05 |
ENSRNOT00000065230
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr16_-_32753278 | 0.05 |
ENSRNOT00000015759
|
Mfap3l
|
microfibrillar-associated protein 3-like |
| chr9_+_16508799 | 0.05 |
ENSRNOT00000021803
|
Rpl7l1
|
ribosomal protein L7-like 1 |
| chr6_-_44363915 | 0.04 |
ENSRNOT00000085925
|
Id2
|
inhibitor of DNA binding 2, HLH protein |
| chr1_+_213451721 | 0.04 |
ENSRNOT00000045652
|
Olr315
|
olfactory receptor 315 |
| chr4_-_170783725 | 0.04 |
ENSRNOT00000072528
|
Wbp11
|
WW domain binding protein 11 |
| chr6_-_110681408 | 0.04 |
ENSRNOT00000078265
|
Angel1
|
angel homolog 1 |
| chr3_-_38090526 | 0.03 |
ENSRNOT00000059430
|
Cacnb4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr16_+_35573058 | 0.03 |
ENSRNOT00000059580
|
Galntl6
|
polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chr10_+_19366793 | 0.03 |
ENSRNOT00000050610
|
Fam196b
|
family with sequence similarity 196, member B |
| chr1_+_199351628 | 0.03 |
ENSRNOT00000078578
|
Bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr1_-_189181901 | 0.03 |
ENSRNOT00000092022
|
Gp2
|
glycoprotein 2 |
| chr14_-_3300200 | 0.03 |
ENSRNOT00000037931
|
Btbd8
|
BTB (POZ) domain containing 8 |
| chr2_+_40554146 | 0.03 |
ENSRNOT00000015138
|
Pde4d
|
phosphodiesterase 4D |
| chr7_+_125576327 | 0.03 |
ENSRNOT00000016679
|
Prr5
|
proline rich 5 |
| chr17_+_30556884 | 0.02 |
ENSRNOT00000080929
ENSRNOT00000022022 |
Eci2
|
enoyl-CoA delta isomerase 2 |
| chr15_+_43905099 | 0.02 |
ENSRNOT00000016568
|
Ebf2
|
early B-cell factor 2 |
| chr3_+_172195844 | 0.02 |
ENSRNOT00000034915
|
Npepl1
|
aminopeptidase-like 1 |
| chr17_+_87274944 | 0.02 |
ENSRNOT00000073718
|
Etl4
|
enhancer trap locus 4 |
| chr3_-_145810834 | 0.02 |
ENSRNOT00000075429
|
LOC102555217
|
zinc finger protein 120-like |
| chr18_+_60392376 | 0.02 |
ENSRNOT00000023890
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr10_+_69737328 | 0.02 |
ENSRNOT00000055999
ENSRNOT00000076773 |
Tmem132e
|
transmembrane protein 132E |
| chr13_+_67611708 | 0.02 |
ENSRNOT00000063833
|
Tpr
|
translocated promoter region, nuclear basket protein |
| chr13_+_49005405 | 0.01 |
ENSRNOT00000092560
ENSRNOT00000076457 |
Lemd1
|
LEM domain containing 1 |
| chr11_+_72705129 | 0.01 |
ENSRNOT00000073330
|
Apod
|
apolipoprotein D |
| chr2_+_38230757 | 0.01 |
ENSRNOT00000048598
|
RGD1564606
|
similar to 60S ribosomal protein L23a |
| chr2_-_210088949 | 0.01 |
ENSRNOT00000070994
|
Rbm15
|
RNA binding motif protein 15 |
| chr16_+_23447366 | 0.01 |
ENSRNOT00000068629
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr8_-_48634797 | 0.01 |
ENSRNOT00000012868
|
Hinfp
|
histone H4 transcription factor |
| chr14_+_110676090 | 0.01 |
ENSRNOT00000029513
|
Fancl
|
Fanconi anemia, complementation group L |
| chr19_-_22632071 | 0.01 |
ENSRNOT00000077275
|
Gpt2
|
glutamic--pyruvic transaminase 2 |
| chr10_+_59725398 | 0.00 |
ENSRNOT00000026156
|
P2rx5
|
purinergic receptor P2X 5 |
| chr18_+_70192493 | 0.00 |
ENSRNOT00000020472
|
Cxxc1
|
CXXC finger protein 1 |
| chr16_-_71237118 | 0.00 |
ENSRNOT00000066974
|
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr2_-_149444548 | 0.00 |
ENSRNOT00000018600
|
P2ry12
|
purinergic receptor P2Y12 |
| chr16_-_21380331 | 0.00 |
ENSRNOT00000015247
|
Atp13a1
|
ATPase 13A1 |
| chr7_-_107391184 | 0.00 |
ENSRNOT00000056793
|
Tmem71
|
transmembrane protein 71 |
| chr7_+_13673934 | 0.00 |
ENSRNOT00000048564
|
Olr1085
|
olfactory receptor 1085 |
| chrX_+_24891156 | 0.00 |
ENSRNOT00000071173
|
Wwc3
|
WWC family member 3 |
| chr1_+_17602281 | 0.00 |
ENSRNOT00000075461
|
LOC103690149
|
jouberin-like |
| chr20_+_3309899 | 0.00 |
ENSRNOT00000001049
|
Abcf1
|
ATP binding cassette subfamily F member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Atoh1_Bhlhe23
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.3 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.2 | GO:1901581 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.1 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.1 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.1 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 | 0.3 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.0 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0001806 | type IV hypersensitivity(GO:0001806) negative regulation of hypersensitivity(GO:0002884) |
| 0.0 | 0.0 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.0 | 0.2 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.3 | GO:0010842 | retina layer formation(GO:0010842) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.0 | 0.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.2 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |