Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
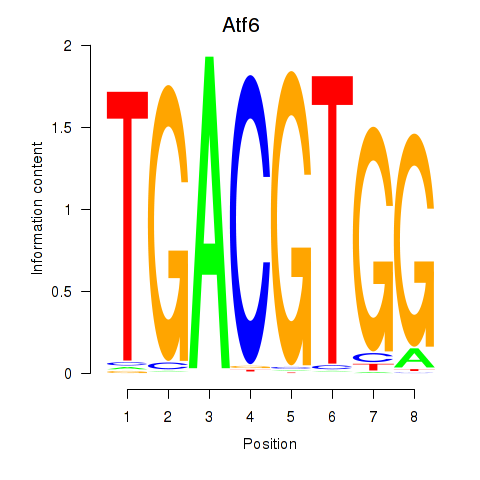
Results for Atf6
Z-value: 0.47
Transcription factors associated with Atf6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Atf6
|
ENSRNOG00000024632 | activating transcription factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atf6 | rn6_v1_chr13_-_89242443_89242443 | 0.11 | 8.6e-01 | Click! |
Activity profile of Atf6 motif
Sorted Z-values of Atf6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_66417741 | 0.38 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr12_-_41671437 | 0.35 |
ENSRNOT00000001883
|
Lhx5
|
LIM homeobox 5 |
| chr1_-_209641123 | 0.19 |
ENSRNOT00000021702
|
Ebf3
|
early B-cell factor 3 |
| chr1_+_274245184 | 0.15 |
ENSRNOT00000018889
|
Dusp5
|
dual specificity phosphatase 5 |
| chr15_+_52241801 | 0.12 |
ENSRNOT00000082639
|
Hr
|
HR, lysine demethylase and nuclear receptor corepressor |
| chr11_+_80255790 | 0.12 |
ENSRNOT00000002522
|
Bcl6
|
B-cell CLL/lymphoma 6 |
| chr10_+_84167331 | 0.11 |
ENSRNOT00000010965
|
Hoxb4
|
homeo box B4 |
| chr19_+_58823814 | 0.10 |
ENSRNOT00000027058
ENSRNOT00000088626 |
Kcnk1
|
potassium two pore domain channel subfamily K member 1 |
| chr11_-_34142753 | 0.08 |
ENSRNOT00000002297
|
Cldn14
|
claudin 14 |
| chr4_-_147756294 | 0.08 |
ENSRNOT00000014537
|
Mbd4
|
methyl-CpG binding domain 4 DNA glycosylase |
| chr11_-_83867203 | 0.07 |
ENSRNOT00000002394
|
Chrd
|
chordin |
| chr1_+_199941161 | 0.07 |
ENSRNOT00000027616
|
Bag3
|
Bcl2-associated athanogene 3 |
| chr9_+_10339075 | 0.07 |
ENSRNOT00000073402
|
Vmac
|
vimentin-type intermediate filament associated coiled-coil protein |
| chr4_-_145390447 | 0.07 |
ENSRNOT00000091965
ENSRNOT00000012136 |
Cidec
|
cell death-inducing DFFA-like effector c |
| chr7_-_11001933 | 0.07 |
ENSRNOT00000084006
|
Tle2
|
transducin-like enhancer of split 2 |
| chr5_+_164845925 | 0.06 |
ENSRNOT00000011384
|
Mthfr
|
methylenetetrahydrofolate reductase |
| chrX_-_63204530 | 0.06 |
ENSRNOT00000076175
|
Zfx
|
zinc finger protein X-linked |
| chr10_+_58784660 | 0.06 |
ENSRNOT00000019925
|
LOC100912483
|
uncharacterized LOC100912483 |
| chr5_+_1417478 | 0.06 |
ENSRNOT00000008153
ENSRNOT00000085564 |
Jph1
|
junctophilin 1 |
| chr2_+_43266739 | 0.06 |
ENSRNOT00000087031
|
Mier3
|
mesoderm induction early response 1, family member 3 |
| chr1_+_78933372 | 0.06 |
ENSRNOT00000029810
|
Ccdc8
|
coiled-coil domain containing 8 |
| chr15_-_109394905 | 0.06 |
ENSRNOT00000019602
ENSRNOT00000078424 |
Tmtc4
|
transmembrane and tetratricopeptide repeat containing 4 |
| chr7_-_58286770 | 0.05 |
ENSRNOT00000005258
|
Rab21
|
RAB21, member RAS oncogene family |
| chr4_+_56711049 | 0.05 |
ENSRNOT00000027237
|
Flnc
|
filamin C |
| chr5_-_147584038 | 0.05 |
ENSRNOT00000010983
|
Zbtb8a
|
zinc finger and BTB domain containing 8a |
| chr10_+_90984227 | 0.05 |
ENSRNOT00000003890
ENSRNOT00000093707 |
Ccdc103
|
coiled-coil domain containing 103 |
| chr12_-_21832813 | 0.05 |
ENSRNOT00000075280
|
Cldn3
|
claudin 3 |
| chr5_-_166726794 | 0.04 |
ENSRNOT00000022799
|
Slc25a33
|
solute carrier family 25 member 33 |
| chr1_+_47162394 | 0.04 |
ENSRNOT00000068457
|
Tmem181
|
transmembrane protein 181 |
| chr3_-_79892429 | 0.04 |
ENSRNOT00000016191
|
Slc39a13
|
solute carrier family 39 member 13 |
| chr10_-_37209881 | 0.04 |
ENSRNOT00000090475
|
Sec24a
|
SEC24 homolog A, COPII coat complex component |
| chr20_+_33527932 | 0.04 |
ENSRNOT00000066492
|
Nepn
|
nephrocan |
| chr4_-_157078130 | 0.04 |
ENSRNOT00000015570
|
Clstn3
|
calsyntenin 3 |
| chr2_+_227455722 | 0.04 |
ENSRNOT00000064809
|
Sec24d
|
SEC24 homolog D, COPII coat complex component |
| chr4_-_152380023 | 0.04 |
ENSRNOT00000012397
|
Erc1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr10_-_96584896 | 0.04 |
ENSRNOT00000004699
ENSRNOT00000055073 |
Prkca
|
protein kinase C, alpha |
| chr6_-_135412312 | 0.04 |
ENSRNOT00000010610
|
Ankrd9
|
ankyrin repeat domain 9 |
| chr7_-_2786856 | 0.04 |
ENSRNOT00000047530
ENSRNOT00000086939 |
Coq10a
|
coenzyme Q10A |
| chr10_+_84135116 | 0.04 |
ENSRNOT00000031035
|
Hoxb7
|
homeo box B7 |
| chrX_+_156999826 | 0.04 |
ENSRNOT00000081819
|
Idh3g
|
isocitrate dehydrogenase 3 (NAD), gamma |
| chr14_+_35683442 | 0.04 |
ENSRNOT00000003085
|
Chic2
|
cysteine-rich hydrophobic domain 2 |
| chr1_-_32643771 | 0.04 |
ENSRNOT00000016975
|
Irx4
|
iroquois homeobox 4 |
| chr16_+_59077574 | 0.04 |
ENSRNOT00000090477
|
Dlc1
|
DLC1 Rho GTPase activating protein |
| chr10_-_102269272 | 0.04 |
ENSRNOT00000048264
|
Cdc42ep4
|
CDC42 effector protein 4 |
| chr2_+_233615739 | 0.04 |
ENSRNOT00000051009
|
Pitx2
|
paired-like homeodomain 2 |
| chr7_-_64341207 | 0.04 |
ENSRNOT00000006051
|
Tmem5
|
transmembrane protein 5 |
| chr2_-_211352540 | 0.04 |
ENSRNOT00000023728
|
LOC108349010
|
protein kish-B |
| chr6_-_80334522 | 0.04 |
ENSRNOT00000059316
|
Fbxo33
|
F-box protein 33 |
| chrX_+_105911925 | 0.04 |
ENSRNOT00000052422
|
LOC108348137
|
armadillo repeat-containing X-linked protein 1 |
| chrX_-_156999650 | 0.04 |
ENSRNOT00000083557
|
Ssr4
|
signal sequence receptor subunit 4 |
| chr16_-_8685529 | 0.04 |
ENSRNOT00000092751
|
Slc18a3
|
solute carrier family 18 member A3 |
| chr1_+_221538104 | 0.04 |
ENSRNOT00000028527
|
Batf2
|
basic leucine zipper ATF-like transcription factor 2 |
| chr6_+_75429729 | 0.04 |
ENSRNOT00000043250
|
Rps10l1
|
ribosomal protein S10-like 1 |
| chr4_+_167754525 | 0.03 |
ENSRNOT00000007889
|
Etv6
|
ets variant 6 |
| chr5_+_76150840 | 0.03 |
ENSRNOT00000020702
|
Dnajc25
|
DnaJ heat shock protein family (Hsp40) member C25 |
| chr2_-_195935878 | 0.03 |
ENSRNOT00000028440
|
Cgn
|
cingulin |
| chr4_-_152380184 | 0.03 |
ENSRNOT00000091473
|
Erc1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr18_-_56534415 | 0.03 |
ENSRNOT00000024374
|
Slc26a2
|
solute carrier family 26 member 2 |
| chr7_+_95309928 | 0.03 |
ENSRNOT00000005887
|
Mtbp
|
MDM2 binding protein |
| chr8_+_129158103 | 0.03 |
ENSRNOT00000025615
|
Eif1b
|
eukaryotic translation initiation factor 1B |
| chr10_-_40764185 | 0.03 |
ENSRNOT00000017486
|
Sparc
|
secreted protein acidic and cysteine rich |
| chr14_+_11198896 | 0.03 |
ENSRNOT00000079767
|
Hnrnpdl
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr7_-_130408187 | 0.03 |
ENSRNOT00000015374
|
Chkb
|
choline kinase beta |
| chr1_-_101086198 | 0.03 |
ENSRNOT00000027917
|
Rcn3
|
reticulocalbin 3 |
| chr6_+_91476698 | 0.03 |
ENSRNOT00000005608
|
Mgat2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr4_+_113948514 | 0.03 |
ENSRNOT00000011521
|
LOC103692171
|
mannosyl-oligosaccharide glucosidase |
| chr4_+_56711275 | 0.03 |
ENSRNOT00000088149
|
Flnc
|
filamin C |
| chr11_+_87224421 | 0.03 |
ENSRNOT00000000308
|
Dgcr14
|
DiGeorge syndrome critical region gene 14 |
| chr9_-_115555261 | 0.03 |
ENSRNOT00000056366
|
Ptprm
|
protein tyrosine phosphatase, receptor type, M |
| chr1_+_167169442 | 0.03 |
ENSRNOT00000048172
|
LOC102549471
|
short transient receptor potential channel 2-like |
| chr6_+_72124417 | 0.03 |
ENSRNOT00000040548
|
Scfd1
|
sec1 family domain containing 1 |
| chr5_+_169452493 | 0.03 |
ENSRNOT00000014444
|
Gpr153
|
G protein-coupled receptor 153 |
| chr9_-_113331319 | 0.03 |
ENSRNOT00000020681
|
Vapa
|
VAMP associated protein A |
| chrX_-_4945944 | 0.03 |
ENSRNOT00000077238
|
Kdm6a
|
lysine demethylase 6A |
| chr14_-_37108341 | 0.03 |
ENSRNOT00000002899
|
Spata18
|
spermatogenesis associated 18 |
| chr1_+_198383201 | 0.03 |
ENSRNOT00000037405
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr11_-_70961358 | 0.03 |
ENSRNOT00000002427
|
Lmln
|
leishmanolysin like peptidase |
| chr12_-_14175945 | 0.03 |
ENSRNOT00000001469
|
Ap5z1
|
adaptor-related protein complex 5, zeta 1 subunit |
| chr14_-_86387606 | 0.02 |
ENSRNOT00000089384
ENSRNOT00000006642 |
Ddx56
|
DEAD-box helicase 56 |
| chr8_-_53146953 | 0.02 |
ENSRNOT00000045356
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr8_-_83421612 | 0.02 |
ENSRNOT00000015824
|
Hcrtr2
|
hypocretin receptor 2 |
| chr5_+_62109328 | 0.02 |
ENSRNOT00000012260
|
Nans
|
N-acetylneuraminate synthase |
| chr17_+_85356042 | 0.02 |
ENSRNOT00000022201
|
Commd3
|
COMM domain containing 3 |
| chr1_-_101085884 | 0.02 |
ENSRNOT00000085352
|
Rcn3
|
reticulocalbin 3 |
| chr7_-_70969905 | 0.02 |
ENSRNOT00000057745
|
Nab2
|
Ngfi-A binding protein 2 |
| chr2_+_124149872 | 0.02 |
ENSRNOT00000023584
|
Spata5
|
spermatogenesis associated 5 |
| chr12_-_24556976 | 0.02 |
ENSRNOT00000041798
|
Bcl7b
|
BCL tumor suppressor 7B |
| chr6_-_88232252 | 0.02 |
ENSRNOT00000047597
|
Rpl10l
|
ribosomal protein L10-like |
| chr1_-_81144024 | 0.02 |
ENSRNOT00000088431
|
Zfp94
|
zinc finger protein 94 |
| chr7_-_24667301 | 0.02 |
ENSRNOT00000009154
|
Tmem263
|
transmembrane protein 263 |
| chr17_+_26925828 | 0.02 |
ENSRNOT00000018310
|
Txndc5
|
thioredoxin domain containing 5 |
| chr13_+_35554964 | 0.02 |
ENSRNOT00000072632
|
Tmem185b
|
transmembrane protein 185B |
| chr12_-_19314016 | 0.02 |
ENSRNOT00000001825
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr5_-_138222534 | 0.02 |
ENSRNOT00000009691
|
Zfp691
|
zinc finger protein 691 |
| chr5_-_62001196 | 0.02 |
ENSRNOT00000012602
|
Trmo
|
tRNA methyltransferase O |
| chr14_-_114484127 | 0.02 |
ENSRNOT00000056706
|
Eml6
|
echinoderm microtubule associated protein like 6 |
| chr10_+_43768708 | 0.02 |
ENSRNOT00000086218
|
Sh3bp5l
|
SH3 binding domain protein 5 like |
| chr1_+_264768105 | 0.02 |
ENSRNOT00000086766
ENSRNOT00000048843 |
Lzts2
|
leucine zipper tumor suppressor 2 |
| chr14_+_1462358 | 0.02 |
ENSRNOT00000077243
|
Csf2ra
|
colony stimulating factor 2 receptor alpha subunit |
| chr10_+_40247436 | 0.02 |
ENSRNOT00000079830
|
Gpx3
|
glutathione peroxidase 3 |
| chr8_-_49075892 | 0.02 |
ENSRNOT00000082687
|
Arcn1
|
archain 1 |
| chr16_+_23668595 | 0.02 |
ENSRNOT00000067886
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr11_-_34598102 | 0.02 |
ENSRNOT00000068743
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr4_+_157554794 | 0.02 |
ENSRNOT00000024116
|
Ing4
|
inhibitor of growth family, member 4 |
| chr12_-_52676608 | 0.02 |
ENSRNOT00000072507
|
Plcxd1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr3_-_161212188 | 0.02 |
ENSRNOT00000065751
|
Wfdc3
|
WAP four-disulfide core domain 3 |
| chr1_-_229639187 | 0.02 |
ENSRNOT00000016804
|
Zfp91
|
zinc finger protein 91 |
| chr13_+_80125391 | 0.02 |
ENSRNOT00000044190
|
Mir199a2
|
microRNA 199a-2 |
| chr13_+_90467265 | 0.02 |
ENSRNOT00000008518
|
Copa
|
coatomer protein complex subunit alpha |
| chr7_+_12144162 | 0.02 |
ENSRNOT00000077122
|
Tcf3
|
transcription factor 3 |
| chr12_+_24473981 | 0.02 |
ENSRNOT00000001973
|
Fzd9
|
frizzled class receptor 9 |
| chr10_+_7279920 | 0.02 |
ENSRNOT00000003749
|
Tmem114
|
transmembrane protein 114 |
| chr16_+_81493355 | 0.02 |
ENSRNOT00000023736
ENSRNOT00000090674 |
Cdc16
|
cell division cycle 16 |
| chr10_+_20591432 | 0.02 |
ENSRNOT00000059780
|
Pank3
|
pantothenate kinase 3 |
| chr5_+_143500441 | 0.02 |
ENSRNOT00000045513
|
Grik3
|
glutamate ionotropic receptor kainate type subunit 3 |
| chr2_+_225854629 | 0.02 |
ENSRNOT00000036931
|
Dnttip2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr17_-_1672021 | 0.02 |
ENSRNOT00000036287
|
Zfp367
|
zinc finger protein 367 |
| chr1_+_220009397 | 0.02 |
ENSRNOT00000084410
|
Rbm4b
|
RNA binding motif protein 4B |
| chr8_+_48422036 | 0.01 |
ENSRNOT00000036051
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr8_-_103554399 | 0.01 |
ENSRNOT00000079965
ENSRNOT00000089833 |
LOC100909709
|
short transient receptor potential channel 1-like |
| chr8_-_67232298 | 0.01 |
ENSRNOT00000049441
|
Spesp1
|
sperm equatorial segment protein 1 |
| chr15_+_61826711 | 0.01 |
ENSRNOT00000068269
|
Elf1
|
E74-like factor 1 |
| chr14_+_44413636 | 0.01 |
ENSRNOT00000003552
|
Smim14
|
small integral membrane protein 14 |
| chr19_-_56731372 | 0.01 |
ENSRNOT00000024182
|
Nup133
|
nucleoporin 133 |
| chr1_+_226947105 | 0.01 |
ENSRNOT00000028373
|
Prpf19
|
pre-mRNA processing factor 19 |
| chr5_-_39650894 | 0.01 |
ENSRNOT00000010421
ENSRNOT00000089522 |
Ufl1
|
Ufm1-specific ligase 1 |
| chr6_-_109205004 | 0.01 |
ENSRNOT00000010512
|
Tmed10
|
transmembrane p24 trafficking protein 10 |
| chr7_+_141249044 | 0.01 |
ENSRNOT00000084911
|
Aqp5
|
aquaporin 5 |
| chr6_+_143901280 | 0.01 |
ENSRNOT00000091884
ENSRNOT00000005648 |
Zfp386
|
zinc finger protein 386 (Kruppel-like) |
| chr3_+_113415774 | 0.01 |
ENSRNOT00000056151
|
Serf2
|
small EDRK-rich factor 2 |
| chr14_+_11199404 | 0.01 |
ENSRNOT00000003106
|
Hnrnpdl
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr2_+_209840404 | 0.01 |
ENSRNOT00000050149
|
Kcna2
|
potassium voltage-gated channel subfamily A member 2 |
| chr5_+_150725654 | 0.01 |
ENSRNOT00000089852
ENSRNOT00000017740 |
Dnajc8
|
DnaJ heat shock protein family (Hsp40) member C8 |
| chr7_+_11295892 | 0.01 |
ENSRNOT00000089004
|
Tjp3
|
tight junction protein 3 |
| chr20_-_48881119 | 0.01 |
ENSRNOT00000018892
|
Qrsl1
|
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr4_-_157486844 | 0.01 |
ENSRNOT00000038281
ENSRNOT00000022874 |
Cops7a
|
COP9 signalosome subunit 7A |
| chr11_-_34598275 | 0.01 |
ENSRNOT00000077233
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chrX_+_73390903 | 0.01 |
ENSRNOT00000077002
|
Zfp449
|
zinc finger protein 449 |
| chr12_+_16340922 | 0.01 |
ENSRNOT00000001696
|
Snx8
|
sorting nexin 8 |
| chr5_+_90800082 | 0.01 |
ENSRNOT00000093696
ENSRNOT00000093206 |
Kdm4c
|
lysine demethylase 4C |
| chr5_-_147784311 | 0.01 |
ENSRNOT00000074172
|
Fam167b
|
family with sequence similarity 167, member B |
| chr10_-_74298599 | 0.01 |
ENSRNOT00000007379
|
Ypel2
|
yippee-like 2 |
| chr2_+_104854572 | 0.01 |
ENSRNOT00000064828
ENSRNOT00000090912 |
Hltf
|
helicase-like transcription factor |
| chr1_-_183729569 | 0.01 |
ENSRNOT00000083293
|
Copb1
|
coatomer protein complex, subunit beta 1 |
| chr12_-_43940798 | 0.01 |
ENSRNOT00000001485
|
RGD1562310
|
similar to hypothetical protein FLJ21415 |
| chr4_-_55713620 | 0.01 |
ENSRNOT00000010246
|
Gcc1
|
GRIP and coiled-coil domain containing 1 |
| chr8_+_117117430 | 0.01 |
ENSRNOT00000073247
|
Gpx1
|
glutathione peroxidase 1 |
| chr1_-_174588529 | 0.01 |
ENSRNOT00000017485
|
Dennd5a
|
DENN domain containing 5A |
| chr16_+_2338333 | 0.01 |
ENSRNOT00000017692
|
Arf4
|
ADP-ribosylation factor 4 |
| chr1_+_162768156 | 0.01 |
ENSRNOT00000049321
|
Pak1
|
p21 (RAC1) activated kinase 1 |
| chr16_+_61758917 | 0.01 |
ENSRNOT00000084858
|
Leprotl1
|
leptin receptor overlapping transcript-like 1 |
| chr10_+_88356615 | 0.01 |
ENSRNOT00000022687
|
Klhl10
|
kelch-like family member 10 |
| chr9_-_61810417 | 0.01 |
ENSRNOT00000020910
|
Rftn2
|
raftlin family member 2 |
| chr19_-_39257451 | 0.01 |
ENSRNOT00000066312
|
Cog8
|
component of oligomeric golgi complex 8 |
| chr10_+_76343847 | 0.01 |
ENSRNOT00000055674
|
Trim25
|
tripartite motif-containing 25 |
| chr11_+_87678295 | 0.01 |
ENSRNOT00000091675
|
Klhl22
|
kelch-like family member 22 |
| chr10_-_90983928 | 0.01 |
ENSRNOT00000055176
|
Eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr1_+_221420271 | 0.01 |
ENSRNOT00000028481
|
LOC687780
|
similar to Finkel-Biskis-Reilly murine sarcoma virusubiquitously expressed |
| chr1_-_32429917 | 0.01 |
ENSRNOT00000024111
|
Lpcat1
|
lysophosphatidylcholine acyltransferase 1 |
| chrX_-_19369746 | 0.01 |
ENSRNOT00000075249
|
LOC108348096
|
ras-related GTP-binding protein B |
| chr1_+_173532803 | 0.01 |
ENSRNOT00000021017
|
Eif3f
|
eukaryotic translation initiation factor 3, subunit F |
| chr4_+_176565694 | 0.01 |
ENSRNOT00000016794
|
Pyroxd1
|
pyridine nucleotide-disulphide oxidoreductase domain 1 |
| chr18_+_58325319 | 0.01 |
ENSRNOT00000065802
ENSRNOT00000077726 |
Napg
|
NSF attachment protein gamma |
| chr10_+_70134729 | 0.01 |
ENSRNOT00000076739
|
Lig3
|
DNA ligase 3 |
| chr3_-_46457201 | 0.01 |
ENSRNOT00000010683
|
Ly75
|
lymphocyte antigen 75 |
| chrX_-_19134869 | 0.01 |
ENSRNOT00000004235
|
RragB
|
Ras-related GTP binding B |
| chr1_-_87174530 | 0.01 |
ENSRNOT00000089578
|
Kcnk6
|
potassium two pore domain channel subfamily K member 6 |
| chr10_+_74436534 | 0.01 |
ENSRNOT00000008298
|
Trim37
|
tripartite motif-containing 37 |
| chr5_+_21632926 | 0.01 |
ENSRNOT00000008522
|
Rab2a
|
RAB2A, member RAS oncogene family |
| chr12_+_23463157 | 0.01 |
ENSRNOT00000044841
|
Cux1
|
cut-like homeobox 1 |
| chr20_+_2501252 | 0.01 |
ENSRNOT00000079307
ENSRNOT00000084559 |
Trim39
|
tripartite motif-containing 39 |
| chr3_+_95232166 | 0.01 |
ENSRNOT00000017952
|
LOC691083
|
hypothetical protein LOC691083 |
| chr1_-_218810118 | 0.01 |
ENSRNOT00000065950
ENSRNOT00000020886 |
Ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr1_-_263718745 | 0.01 |
ENSRNOT00000074810
|
Dnmbp
|
dynamin binding protein |
| chr7_+_114724610 | 0.01 |
ENSRNOT00000014541
|
Dennd3
|
DENN domain containing 3 |
| chrX_-_1856258 | 0.01 |
ENSRNOT00000079980
|
RGD1564855
|
similar to RuvB-like protein 1 |
| chr15_+_61826937 | 0.01 |
ENSRNOT00000084005
ENSRNOT00000079417 |
Elf1
|
E74-like factor 1 |
| chr8_-_25904564 | 0.01 |
ENSRNOT00000082744
ENSRNOT00000064783 |
Tbx20
|
T-box 20 |
| chr1_-_205842428 | 0.01 |
ENSRNOT00000024655
|
Dhx32
|
DEAH-box helicase 32 (putative) |
| chr6_+_52459946 | 0.00 |
ENSRNOT00000073277
ENSRNOT00000013474 |
Atxn7l1
|
ataxin 7-like 1 |
| chr10_+_15241590 | 0.00 |
ENSRNOT00000037372
ENSRNOT00000037381 |
Mettl26
|
methyltransferase like 26 |
| chr2_+_144591755 | 0.00 |
ENSRNOT00000044424
|
Sohlh2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr1_+_77994203 | 0.00 |
ENSRNOT00000002044
|
Napa
|
NSF attachment protein alpha |
| chr1_+_224939497 | 0.00 |
ENSRNOT00000025664
|
Stx5
|
syntaxin 5 |
| chr1_-_175895510 | 0.00 |
ENSRNOT00000064535
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr6_-_86713370 | 0.00 |
ENSRNOT00000005821
|
Klhl28
|
kelch-like family member 28 |
| chr1_-_100865894 | 0.00 |
ENSRNOT00000027580
|
Ptov1
|
prostate tumor overexpressed 1 |
| chr6_-_94818349 | 0.00 |
ENSRNOT00000005933
|
Gpr135
|
G protein-coupled receptor 135 |
| chr11_+_77239098 | 0.00 |
ENSRNOT00000058701
|
Gmnc
|
geminin coiled-coil domain containing |
| chr1_+_282188110 | 0.00 |
ENSRNOT00000013845
|
Fam45a
|
family with sequence similarity 45, member A |
| chr2_+_207108552 | 0.00 |
ENSRNOT00000027234
|
Slc16a1
|
solute carrier family 16 member 1 |
| chr20_-_11737050 | 0.00 |
ENSRNOT00000001637
|
Sumo3
|
small ubiquitin-like modifier 3 |
| chr4_+_56625561 | 0.00 |
ENSRNOT00000008356
ENSRNOT00000090808 ENSRNOT00000008972 |
Calu
|
calumenin |
| chr10_+_74436208 | 0.00 |
ENSRNOT00000090921
ENSRNOT00000091163 |
Trim37
|
tripartite motif-containing 37 |
| chr3_+_161212156 | 0.00 |
ENSRNOT00000020280
ENSRNOT00000083553 |
Dnttip1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr4_-_62663200 | 0.00 |
ENSRNOT00000040969
|
Cnot4
|
CCR4-NOT transcription complex, subunit 4 |
| chr12_-_48663143 | 0.00 |
ENSRNOT00000082809
|
Ficd
|
FIC domain containing |
| chr10_-_84789832 | 0.00 |
ENSRNOT00000071719
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr7_-_140506925 | 0.00 |
ENSRNOT00000083354
|
Kmt2d
|
lysine methyltransferase 2D |
| chr20_+_2004052 | 0.00 |
ENSRNOT00000001008
|
Mog
|
myelin oligodendrocyte glycoprotein |
Network of associatons between targets according to the STRING database.
First level regulatory network of Atf6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.4 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.0 | 0.1 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) regulation of memory T cell differentiation(GO:0043380) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.0 | GO:0060577 | pulmonary vein morphogenesis(GO:0060577) |
| 0.0 | 0.0 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.0 | 0.1 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.0 | 0.0 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.0 | GO:0005962 | mitochondrial isocitrate dehydrogenase complex (NAD+)(GO:0005962) isocitrate dehydrogenase complex (NAD+)(GO:0045242) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.0 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.0 | 0.0 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.1 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |