Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
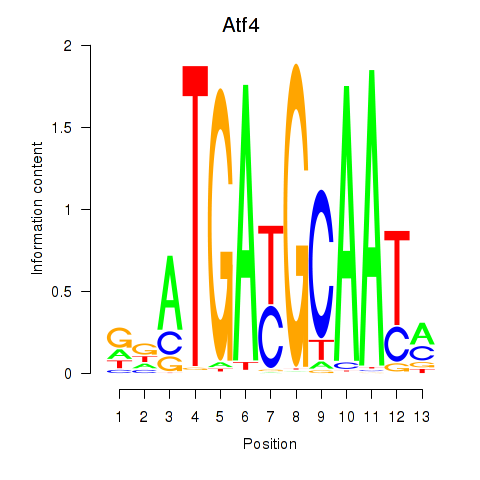
Results for Atf4
Z-value: 0.38
Transcription factors associated with Atf4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Atf4
|
ENSRNOG00000017801 | activating transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atf4 | rn6_v1_chr7_+_121480723_121480723 | 0.74 | 1.5e-01 | Click! |
Activity profile of Atf4 motif
Sorted Z-values of Atf4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_78969013 | 0.15 |
ENSRNOT00000019772
ENSRNOT00000057585 |
Fn1
|
fibronectin 1 |
| chr6_-_125723944 | 0.15 |
ENSRNOT00000007004
|
Fbln5
|
fibulin 5 |
| chr19_-_11057254 | 0.14 |
ENSRNOT00000025559
|
Herpud1
|
homocysteine inducible ER protein with ubiquitin like domain 1 |
| chr5_-_137372993 | 0.14 |
ENSRNOT00000092823
|
Tmem125
|
transmembrane protein 125 |
| chr19_+_14508616 | 0.13 |
ENSRNOT00000019192
|
Hmox1
|
heme oxygenase 1 |
| chr5_+_136669674 | 0.13 |
ENSRNOT00000048770
|
Slc6a9
|
solute carrier family 6 member 9 |
| chr6_-_125723732 | 0.12 |
ENSRNOT00000084815
|
Fbln5
|
fibulin 5 |
| chr7_+_130296897 | 0.10 |
ENSRNOT00000044854
|
Adm2
|
adrenomedullin 2 |
| chr15_-_33527031 | 0.10 |
ENSRNOT00000019979
|
Homez
|
homeobox and leucine zipper encoding |
| chr1_-_101596822 | 0.10 |
ENSRNOT00000028490
|
Fgf21
|
fibroblast growth factor 21 |
| chr2_-_149347796 | 0.08 |
ENSRNOT00000091020
|
P2ry14
|
purinergic receptor P2Y14 |
| chr7_-_101140308 | 0.07 |
ENSRNOT00000006279
|
Fam84b
|
family with sequence similarity 84, member B |
| chr3_+_111160205 | 0.07 |
ENSRNOT00000019392
|
Chac1
|
ChaC glutathione-specific gamma-glutamylcyclotransferase 1 |
| chr10_-_38782419 | 0.07 |
ENSRNOT00000073964
|
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr7_+_70580198 | 0.06 |
ENSRNOT00000083472
ENSRNOT00000008941 |
Ddit3
|
DNA-damage inducible transcript 3 |
| chr10_-_20622600 | 0.06 |
ENSRNOT00000020554
|
Fbll1
|
fibrillarin-like 1 |
| chr19_+_53629779 | 0.06 |
ENSRNOT00000051352
|
Map1lc3b
|
microtubule-associated protein 1 light chain 3 beta |
| chr1_+_79727597 | 0.06 |
ENSRNOT00000034790
|
AC093995.1
|
|
| chr15_-_6587367 | 0.06 |
ENSRNOT00000038449
|
Zfp385d
|
zinc finger protein 385D |
| chr4_-_44136815 | 0.06 |
ENSRNOT00000086810
|
Tfec
|
transcription factor EC |
| chr8_+_22856539 | 0.06 |
ENSRNOT00000015381
|
Angptl8
|
angiopoietin-like 8 |
| chr4_-_157829241 | 0.06 |
ENSRNOT00000026184
|
Ltbr
|
lymphotoxin beta receptor |
| chr2_-_227411964 | 0.05 |
ENSRNOT00000019931
|
Synpo2
|
synaptopodin 2 |
| chr19_+_37668133 | 0.05 |
ENSRNOT00000044971
|
Pard6a
|
par-6 family cell polarity regulator alpha |
| chr19_-_43215281 | 0.05 |
ENSRNOT00000025052
|
Aars
|
alanyl-tRNA synthetase |
| chr1_+_100473643 | 0.05 |
ENSRNOT00000026379
|
Josd2
|
Josephin domain containing 2 |
| chr19_-_43215077 | 0.05 |
ENSRNOT00000082151
|
Aars
|
alanyl-tRNA synthetase |
| chr4_-_115015965 | 0.04 |
ENSRNOT00000014603
|
Mthfd2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr5_+_145079803 | 0.04 |
ENSRNOT00000084202
|
Sfpq
|
splicing factor proline and glutamine rich |
| chr9_+_88110731 | 0.04 |
ENSRNOT00000088677
|
Rhbdd1
|
rhomboid domain containing 1 |
| chr16_-_71040847 | 0.04 |
ENSRNOT00000020606
|
Star
|
steroidogenic acute regulatory protein |
| chr7_-_141185710 | 0.04 |
ENSRNOT00000085033
|
Faim2
|
Fas apoptotic inhibitory molecule 2 |
| chr12_-_11240101 | 0.04 |
ENSRNOT00000076423
|
Bud31
|
BUD31 homolog |
| chr18_-_60002529 | 0.04 |
ENSRNOT00000081014
ENSRNOT00000024264 ENSRNOT00000059162 |
Nars
|
asparaginyl-tRNA synthetase |
| chr5_+_147375350 | 0.04 |
ENSRNOT00000010674
|
Yars
|
tyrosyl-tRNA synthetase |
| chr4_+_85235172 | 0.03 |
ENSRNOT00000014780
|
Gars
|
glycyl-tRNA synthetase |
| chr19_+_37668693 | 0.03 |
ENSRNOT00000084970
|
Pard6a
|
par-6 family cell polarity regulator alpha |
| chr1_-_216801656 | 0.03 |
ENSRNOT00000050721
|
Cars
|
cysteinyl-tRNA synthetase |
| chr10_-_75120247 | 0.03 |
ENSRNOT00000011402
|
Lpo
|
lactoperoxidase |
| chr5_+_62071346 | 0.03 |
ENSRNOT00000012478
|
Anp32b
|
acidic nuclear phosphoprotein 32 family member B |
| chr1_-_194769524 | 0.03 |
ENSRNOT00000025988
|
Nupr1
|
nuclear protein 1, transcriptional regulator |
| chr9_+_74124016 | 0.03 |
ENSRNOT00000019023
|
Cps1
|
carbamoyl-phosphate synthase 1 |
| chr1_+_273854248 | 0.02 |
ENSRNOT00000044827
ENSRNOT00000017600 ENSRNOT00000086496 |
Add3
|
adducin 3 |
| chr7_-_139063752 | 0.02 |
ENSRNOT00000072309
|
LOC102551901
|
protein lifeguard 2-like |
| chr1_-_198045154 | 0.02 |
ENSRNOT00000072875
|
Nupr1l1
|
nuclear protein, transcriptional regulator, 1-like 1 |
| chr6_+_109466060 | 0.02 |
ENSRNOT00000011339
|
Jdp2
|
Jun dimerization protein 2 |
| chr2_-_189333322 | 0.02 |
ENSRNOT00000073599
ENSRNOT00000071253 ENSRNOT00000076325 |
Hax1
|
HCLS1 associated protein X-1 |
| chr5_+_128450680 | 0.02 |
ENSRNOT00000010700
|
Txndc12
|
thioredoxin domain containing 12 |
| chr8_+_118013612 | 0.02 |
ENSRNOT00000056166
|
Map4
|
microtubule-associated protein 4 |
| chr12_-_11240383 | 0.02 |
ENSRNOT00000076149
ENSRNOT00000075987 ENSRNOT00000001307 |
Bud31
|
BUD31 homolog |
| chr16_-_70998575 | 0.02 |
ENSRNOT00000019935
|
Kcnu1
|
potassium calcium-activated channel subfamily U member 1 |
| chr10_+_84119884 | 0.02 |
ENSRNOT00000009951
|
Hoxb9
|
homeo box B9 |
| chr5_-_150704117 | 0.02 |
ENSRNOT00000067455
|
Sesn2
|
sestrin 2 |
| chr14_+_44889287 | 0.02 |
ENSRNOT00000091312
ENSRNOT00000032273 |
Tmem156
|
transmembrane protein 156 |
| chr18_-_67224566 | 0.01 |
ENSRNOT00000064947
|
Dcc
|
DCC netrin 1 receptor |
| chr1_-_263269762 | 0.01 |
ENSRNOT00000022309
|
Got1
|
glutamic-oxaloacetic transaminase 1 |
| chr15_-_35113678 | 0.01 |
ENSRNOT00000059677
|
Ctsg
|
cathepsin G |
| chr4_-_163261958 | 0.01 |
ENSRNOT00000086390
|
Olr1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr19_+_49695579 | 0.01 |
ENSRNOT00000016963
|
Gan
|
gigaxonin |
| chr1_+_102849889 | 0.01 |
ENSRNOT00000066791
|
Gtf2h1
|
general transcription factor IIH subunit 1 |
| chr7_-_70829815 | 0.01 |
ENSRNOT00000011082
|
Shmt2
|
serine hydroxymethyltransferase 2 |
| chr4_+_145440114 | 0.01 |
ENSRNOT00000013052
|
Creld1
|
cysteine-rich with EGF-like domains 1 |
| chr7_-_70643169 | 0.01 |
ENSRNOT00000010106
|
Inhbe
|
inhibin beta E subunit |
| chr1_-_199624783 | 0.01 |
ENSRNOT00000026908
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chr7_+_141054494 | 0.01 |
ENSRNOT00000088176
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr18_-_36579403 | 0.01 |
ENSRNOT00000025284
|
Lars
|
leucyl-tRNA synthetase |
| chr7_-_117773134 | 0.01 |
ENSRNOT00000045135
|
Recql4
|
RecQ like helicase 4 |
| chr4_+_33638709 | 0.00 |
ENSRNOT00000009888
ENSRNOT00000034719 ENSRNOT00000052333 |
Tac1
|
tachykinin, precursor 1 |
| chr9_-_84383687 | 0.00 |
ENSRNOT00000019685
|
Farsb
|
phenylalanyl-tRNA synthetase, beta subunit |
| chr12_+_11240761 | 0.00 |
ENSRNOT00000001310
|
Pdap1
|
PDGFA associated protein 1 |
| chr1_+_142050458 | 0.00 |
ENSRNOT00000018107
|
Ngrn
|
neugrin, neurite outgrowth associated |
| chr8_-_13680870 | 0.00 |
ENSRNOT00000081616
|
Hephl1
|
hephaestin-like 1 |
| chr3_+_170399302 | 0.00 |
ENSRNOT00000035245
|
Cass4
|
Cas scaffolding protein family member 4 |
| chr11_+_80319177 | 0.00 |
ENSRNOT00000036777
|
Rtp2
|
receptor (chemosensory) transporter protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Atf4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.0 | 0.1 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.0 | 0.1 | GO:1904640 | response to methionine(GO:1904640) |
| 0.0 | 0.1 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.0 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.0 | GO:0017143 | insecticide metabolic process(GO:0017143) |
| 0.0 | 0.0 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.1 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.0 | GO:0015960 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.0 | 0.1 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:1990622 | CHOP-C/EBP complex(GO:0036488) CHOP-ATF3 complex(GO:1990622) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.2 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.0 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |