Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
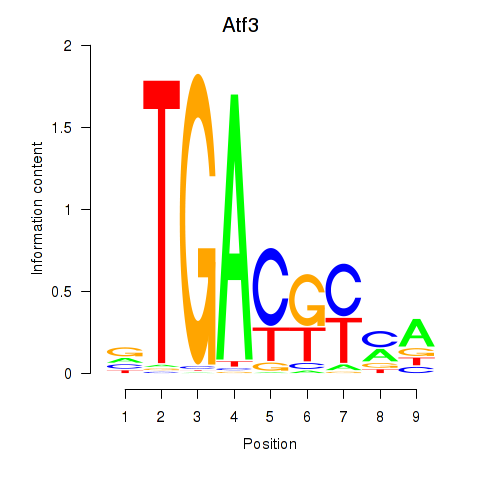
Results for Atf3
Z-value: 0.83
Transcription factors associated with Atf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Atf3
|
ENSRNOG00000003745 | activating transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atf3 | rn6_v1_chr13_-_109844359_109844359 | -0.75 | 1.5e-01 | Click! |
Activity profile of Atf3 motif
Sorted Z-values of Atf3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_55696601 | 0.37 |
ENSRNOT00000016127
|
RGD1562914
|
RGD1562914 |
| chr3_+_114355798 | 0.34 |
ENSRNOT00000024658
ENSRNOT00000036435 |
Slc28a2
|
solute carrier family 28 member 2 |
| chrX_+_15098904 | 0.30 |
ENSRNOT00000007367
ENSRNOT00000087033 |
Rbm3
|
RNA binding motif (RNP1, RRM) protein 3 |
| chr20_-_3793985 | 0.29 |
ENSRNOT00000049540
ENSRNOT00000086293 |
RT1-CE16
|
RT1 class I, locus CE16 |
| chrX_-_70460536 | 0.26 |
ENSRNOT00000076824
|
Pdzd11
|
PDZ domain containing 11 |
| chr2_+_123734199 | 0.26 |
ENSRNOT00000038729
|
RGD1307100
|
similar to RIKEN cDNA D630029K19 |
| chr4_+_123801174 | 0.25 |
ENSRNOT00000029055
|
RGD1560289
|
similar to chromosome 3 open reading frame 20 |
| chr4_+_51614676 | 0.25 |
ENSRNOT00000060494
|
Asb15
|
ankyrin repeat and SOCS box containing 15 |
| chr5_+_131719922 | 0.23 |
ENSRNOT00000010524
|
Spata6
|
spermatogenesis associated 6 |
| chr6_+_127941526 | 0.23 |
ENSRNOT00000033897
|
LOC500712
|
Ab1-233 |
| chr10_+_82775691 | 0.23 |
ENSRNOT00000030737
|
Hils1
|
histone linker H1 domain, spermatid-specific 1 |
| chr19_+_55300395 | 0.22 |
ENSRNOT00000092169
|
Ctu2
|
cytosolic thiouridylase subunit 2 |
| chr9_+_49903085 | 0.21 |
ENSRNOT00000022601
|
RGD1310553
|
similar to expressed sequence AI597479 |
| chr2_+_30664639 | 0.21 |
ENSRNOT00000076372
ENSRNOT00000076294 ENSRNOT00000076434 ENSRNOT00000076484 |
Taf9
|
TATA-box binding protein associated factor 9 |
| chr17_-_1797732 | 0.21 |
ENSRNOT00000033017
ENSRNOT00000079742 |
Cdc14b
|
cell division cycle 14B |
| chr1_-_37886675 | 0.20 |
ENSRNOT00000024149
|
RGD1566325
|
similar to regulator of sex-limitation candidate 16 |
| chr10_-_47453442 | 0.20 |
ENSRNOT00000050061
|
Usp22
|
ubiquitin specific peptidase 22 |
| chr8_+_117737387 | 0.20 |
ENSRNOT00000090164
ENSRNOT00000091573 |
Pfkfb4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr9_-_120101098 | 0.20 |
ENSRNOT00000002917
|
Fam114a1l1
|
family with sequence similarity 114, member A1-like 1 |
| chr14_-_17582823 | 0.20 |
ENSRNOT00000087686
|
LOC100911949
|
uncharacterized LOC100911949 |
| chr1_-_219422268 | 0.20 |
ENSRNOT00000025092
|
Carns1
|
carnosine synthase 1 |
| chr3_-_2459385 | 0.20 |
ENSRNOT00000014080
|
Cysrt1
|
cysteine rich tail 1 |
| chr17_-_10622226 | 0.18 |
ENSRNOT00000044559
|
Simc1
|
SUMO-interacting motifs containing 1 |
| chr2_+_155555840 | 0.18 |
ENSRNOT00000080951
|
AABR07010944.1
|
|
| chr15_+_10462749 | 0.18 |
ENSRNOT00000008035
|
Oxsm
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr10_+_57239993 | 0.18 |
ENSRNOT00000067919
|
LOC687707
|
hypothetical protein LOC687707 |
| chr2_+_266141581 | 0.17 |
ENSRNOT00000078187
ENSRNOT00000051951 |
Rpe65
|
RPE65, retinoid isomerohydrolase |
| chr19_-_54652381 | 0.17 |
ENSRNOT00000065472
|
Klhdc4
|
kelch domain containing 4 |
| chr19_+_37990374 | 0.17 |
ENSRNOT00000026827
|
Dus2
|
dihydrouridine synthase 2 |
| chr8_-_49502647 | 0.17 |
ENSRNOT00000040313
|
Tmprss4
|
transmembrane protease, serine 4 |
| chr11_-_62451149 | 0.16 |
ENSRNOT00000093686
ENSRNOT00000081443 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr10_+_5352933 | 0.16 |
ENSRNOT00000003465
|
Tekt5
|
tektin 5 |
| chr14_+_63405408 | 0.16 |
ENSRNOT00000086658
|
AABR07015559.1
|
|
| chr9_-_5330815 | 0.16 |
ENSRNOT00000014548
|
Slc5a7
|
solute carrier family 5 member 7 |
| chr19_-_41567013 | 0.16 |
ENSRNOT00000022757
|
Zfp612
|
zinc finger protein 612 |
| chr2_+_3662763 | 0.16 |
ENSRNOT00000017828
|
Mctp1
|
multiple C2 and transmembrane domain containing 1 |
| chr1_+_98228573 | 0.15 |
ENSRNOT00000019278
|
LOC102555672
|
zinc finger protein 850-like |
| chr4_+_88832178 | 0.15 |
ENSRNOT00000088983
|
Abcg2
|
ATP-binding cassette, subfamily G (WHITE), member 2 |
| chr13_-_113817995 | 0.15 |
ENSRNOT00000057151
|
Cd46
|
CD46 molecule |
| chr4_-_161743847 | 0.15 |
ENSRNOT00000064174
ENSRNOT00000084173 ENSRNOT00000064215 |
Itfg2
|
integrin alpha FG-GAP repeat containing 2 |
| chr11_+_60102121 | 0.15 |
ENSRNOT00000045521
|
Tmprss7
|
transmembrane protease, serine 7 |
| chr4_-_145487426 | 0.15 |
ENSRNOT00000013488
|
Emc3
|
ER membrane protein complex subunit 3 |
| chr4_-_155740193 | 0.15 |
ENSRNOT00000043229
|
LOC102550396
|
LRRGT00188 |
| chr4_+_25635765 | 0.15 |
ENSRNOT00000009340
|
Gtpbp10
|
GTP binding protein 10 |
| chr14_+_115166416 | 0.15 |
ENSRNOT00000088916
ENSRNOT00000078329 |
Psme4
|
proteasome activator subunit 4 |
| chr12_-_41266430 | 0.15 |
ENSRNOT00000001853
ENSRNOT00000081108 |
Oas1b
|
2-5 oligoadenylate synthetase 1B |
| chr6_+_144156175 | 0.14 |
ENSRNOT00000006582
|
Esyt2
|
extended synaptotagmin 2 |
| chr20_-_45815940 | 0.14 |
ENSRNOT00000073276
|
Gpr6
|
G protein-coupled receptor 6 |
| chr14_-_44767120 | 0.14 |
ENSRNOT00000003991
|
Wdr19
|
WD repeat domain 19 |
| chr15_+_24159647 | 0.14 |
ENSRNOT00000082675
|
Lgals3
|
galectin 3 |
| chr1_+_228684136 | 0.14 |
ENSRNOT00000028608
|
Olr337
|
olfactory receptor 337 |
| chr10_-_66602987 | 0.14 |
ENSRNOT00000017949
|
Wsb1
|
WD repeat and SOCS box-containing 1 |
| chr6_+_64224861 | 0.14 |
ENSRNOT00000093159
ENSRNOT00000093664 |
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr8_+_72405748 | 0.14 |
ENSRNOT00000023952
|
Car12
|
carbonic anhydrase 12 |
| chr15_-_47442664 | 0.14 |
ENSRNOT00000072994
|
Prss55
|
protease, serine, 55 |
| chr16_-_21473808 | 0.14 |
ENSRNOT00000066776
|
Zfp964
|
zinc finger protein 964 |
| chr13_-_47397890 | 0.14 |
ENSRNOT00000005505
|
C4bpb
|
complement component 4 binding protein, beta |
| chr1_-_213534641 | 0.14 |
ENSRNOT00000033074
|
Syce1
|
synaptonemal complex central element protein 1 |
| chr11_+_82848853 | 0.14 |
ENSRNOT00000073743
|
Thpo
|
thrombopoietin |
| chr1_-_206417143 | 0.14 |
ENSRNOT00000091863
ENSRNOT00000073836 |
LOC100302465
|
hypothetical LOC100302465 |
| chr4_-_150520774 | 0.14 |
ENSRNOT00000009095
|
Zfp9
|
zinc finger protein 9 |
| chr18_-_61788859 | 0.13 |
ENSRNOT00000034075
ENSRNOT00000034069 |
Ccbe1
|
collagen and calcium binding EGF domains 1 |
| chr10_-_97582188 | 0.13 |
ENSRNOT00000005076
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr20_+_4967194 | 0.13 |
ENSRNOT00000070846
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr2_+_95077577 | 0.13 |
ENSRNOT00000046368
|
Mrps28
|
mitochondrial ribosomal protein S28 |
| chr1_-_219412816 | 0.13 |
ENSRNOT00000083204
ENSRNOT00000029580 |
Rps6kb2
|
ribosomal protein S6 kinase B2 |
| chr6_-_4520604 | 0.13 |
ENSRNOT00000042230
ENSRNOT00000043870 ENSRNOT00000070918 ENSRNOT00000046246 ENSRNOT00000052367 ENSRNOT00000042251 |
Slc8a1
|
solute carrier family 8 member A1 |
| chr10_-_1744647 | 0.13 |
ENSRNOT00000081886
|
AABR07028997.1
|
|
| chr5_-_105579959 | 0.13 |
ENSRNOT00000010827
|
Slc24a2
|
solute carrier family 24 member 2 |
| chr14_+_113530470 | 0.13 |
ENSRNOT00000004919
|
Pnpt1
|
polyribonucleotide nucleotidyltransferase 1 |
| chr1_-_65681440 | 0.13 |
ENSRNOT00000026305
|
Zfp128
|
zinc finger protein 128 |
| chr1_-_128695995 | 0.13 |
ENSRNOT00000077020
|
Synm
|
synemin |
| chr10_+_11373346 | 0.13 |
ENSRNOT00000044624
|
LOC108348151
|
coronin-7-like |
| chr7_+_130542202 | 0.13 |
ENSRNOT00000079501
ENSRNOT00000045647 |
Acr
|
acrosin |
| chr15_-_7400421 | 0.13 |
ENSRNOT00000082997
|
AABR07017045.1
|
|
| chr17_+_11683862 | 0.13 |
ENSRNOT00000024766
|
Msx2
|
msh homeobox 2 |
| chr13_+_96303703 | 0.13 |
ENSRNOT00000084718
ENSRNOT00000029723 |
Efcab2
|
EF-hand calcium binding domain 2 |
| chr1_-_254735548 | 0.13 |
ENSRNOT00000025258
|
Ankrd1
|
ankyrin repeat domain 1 |
| chr1_+_84009268 | 0.13 |
ENSRNOT00000057230
ENSRNOT00000081121 |
RGD1560854
|
similar to FLJ41131 protein |
| chr1_-_219745654 | 0.12 |
ENSRNOT00000054848
|
LOC689065
|
hypothetical protein LOC689065 |
| chr9_+_19451630 | 0.12 |
ENSRNOT00000065048
|
Enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr1_+_88581206 | 0.12 |
ENSRNOT00000087931
|
Zfp382
|
zinc finger protein 382 |
| chr1_+_31967978 | 0.12 |
ENSRNOT00000081471
ENSRNOT00000021532 |
Trip13
|
thyroid hormone receptor interactor 13 |
| chr7_+_144647587 | 0.12 |
ENSRNOT00000022398
|
Hoxc4
|
homeo box C4 |
| chr8_+_29714285 | 0.12 |
ENSRNOT00000042890
|
Opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr7_-_49250953 | 0.12 |
ENSRNOT00000066975
ENSRNOT00000082141 |
Acss3
|
acyl-CoA synthetase short-chain family member 3 |
| chrX_-_23246719 | 0.12 |
ENSRNOT00000085588
|
AABR07037536.1
|
|
| chr9_+_20004280 | 0.12 |
ENSRNOT00000075670
|
Ankrd66
|
ankyrin repeat domain 66 |
| chr12_+_16170162 | 0.12 |
ENSRNOT00000001686
|
Grifin
|
galectin-related inter-fiber protein |
| chr12_-_37185548 | 0.12 |
ENSRNOT00000001338
|
LOC102556092
|
uncharacterized LOC102556092 |
| chr11_-_82938357 | 0.12 |
ENSRNOT00000035945
|
Map3k13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr16_-_25192675 | 0.12 |
ENSRNOT00000032289
|
March1
|
membrane associated ring-CH-type finger 1 |
| chr5_-_139933764 | 0.12 |
ENSRNOT00000015278
|
LOC680700
|
similar to ribosomal protein L10a |
| chrX_-_54303729 | 0.12 |
ENSRNOT00000087919
ENSRNOT00000064340 ENSRNOT00000051249 ENSRNOT00000087547 |
Gk
|
glycerol kinase |
| chr10_-_47018537 | 0.12 |
ENSRNOT00000068351
ENSRNOT00000080083 |
Top3a
|
topoisomerase (DNA) III alpha |
| chr5_-_21345805 | 0.12 |
ENSRNOT00000081296
ENSRNOT00000007802 |
Car8
|
carbonic anhydrase 8 |
| chr7_-_135800481 | 0.12 |
ENSRNOT00000029097
|
Pus7l
|
pseudouridylate synthase 7-like |
| chr15_-_18096039 | 0.12 |
ENSRNOT00000042545
|
Ptgdr
|
prostaglandin D2 receptor |
| chr20_+_28027054 | 0.12 |
ENSRNOT00000071386
ENSRNOT00000001044 |
Ranbp2
|
RAN binding protein 2 |
| chr12_-_10391270 | 0.12 |
ENSRNOT00000092340
|
Wasf3
|
WAS protein family, member 3 |
| chr17_+_66548818 | 0.12 |
ENSRNOT00000024361
|
Anlnl1
|
anillin, actin binding protein-like 1 |
| chr6_-_50941248 | 0.11 |
ENSRNOT00000084533
|
Dus4l
|
dihydrouridine synthase 4-like |
| chr3_+_11811962 | 0.11 |
ENSRNOT00000060258
ENSRNOT00000039151 |
Ptrh1
|
peptidyl-tRNA hydrolase 1 homolog |
| chr8_+_117737117 | 0.11 |
ENSRNOT00000028039
|
Pfkfb4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr16_-_74072541 | 0.11 |
ENSRNOT00000089722
|
AABR07026361.2
|
|
| chr1_-_206394346 | 0.11 |
ENSRNOT00000038122
|
LOC100302465
|
hypothetical LOC100302465 |
| chrM_+_9870 | 0.11 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr1_-_277902279 | 0.11 |
ENSRNOT00000078416
|
Ablim1
|
actin-binding LIM protein 1 |
| chr8_-_39201588 | 0.11 |
ENSRNOT00000011226
|
Stt3a
|
STT3A, catalytic subunit of the oligosaccharyltransferase complex |
| chr4_-_67520356 | 0.11 |
ENSRNOT00000014604
|
Braf
|
B-Raf proto-oncogene, serine/threonine kinase |
| chr18_+_1142782 | 0.11 |
ENSRNOT00000046718
|
Thoc1
|
THO complex 1 |
| chrX_-_31780425 | 0.11 |
ENSRNOT00000004693
|
Asb9
|
ankyrin repeat and SOCS box-containing 9 |
| chr13_+_74456487 | 0.11 |
ENSRNOT00000065801
|
Angptl1
|
angiopoietin-like 1 |
| chr12_+_40895515 | 0.11 |
ENSRNOT00000046323
|
Ptpn11
|
protein tyrosine phosphatase, non-receptor type 11 |
| chr1_-_167308827 | 0.11 |
ENSRNOT00000027590
ENSRNOT00000027575 |
Nup98
|
nucleoporin 98 |
| chr19_+_43163129 | 0.11 |
ENSRNOT00000073721
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr5_-_169658875 | 0.11 |
ENSRNOT00000015840
|
Kcnab2
|
potassium voltage-gated channel subfamily A regulatory beta subunit 2 |
| chr20_+_2194709 | 0.11 |
ENSRNOT00000001017
|
Trim15
|
tripartite motif containing 15 |
| chr13_+_51534025 | 0.11 |
ENSRNOT00000006637
|
Syt2
|
synaptotagmin 2 |
| chr7_+_15785410 | 0.11 |
ENSRNOT00000082664
ENSRNOT00000073235 |
Zfp955a
|
zinc finger protein 955A |
| chr1_-_242861767 | 0.11 |
ENSRNOT00000021185
|
Cbwd1
|
COBW domain containing 1 |
| chr7_-_141624972 | 0.10 |
ENSRNOT00000078785
|
AABR07058884.1
|
|
| chr5_+_126812187 | 0.10 |
ENSRNOT00000057372
|
Ldlrad1
|
low density lipoprotein receptor class A domain containing 1 |
| chr6_+_28515025 | 0.10 |
ENSRNOT00000088033
ENSRNOT00000005317 ENSRNOT00000081141 |
Dnajc27
|
DnaJ heat shock protein family (Hsp40) member C27 |
| chr4_+_70572942 | 0.10 |
ENSRNOT00000051964
|
AC142181.1
|
|
| chr8_-_23390773 | 0.10 |
ENSRNOT00000019601
ENSRNOT00000071846 |
Anln
|
anillin, actin binding protein |
| chr16_-_19097329 | 0.10 |
ENSRNOT00000030617
|
RGD1311847
|
similar to 1700030K09Rik protein |
| chr4_+_152450126 | 0.10 |
ENSRNOT00000076930
|
Rad52
|
RAD52 homolog, DNA repair protein |
| chr2_-_23289266 | 0.10 |
ENSRNOT00000061708
|
Bhmt2
|
betaine-homocysteine S-methyltransferase 2 |
| chr14_-_80130139 | 0.10 |
ENSRNOT00000091652
ENSRNOT00000010482 |
Ablim2
|
actin binding LIM protein family, member 2 |
| chr10_+_83081168 | 0.10 |
ENSRNOT00000035023
|
Tac4
|
tachykinin 4 (hemokinin) |
| chr4_+_140377565 | 0.10 |
ENSRNOT00000082723
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr9_-_81868086 | 0.10 |
ENSRNOT00000067080
|
Zfp142
|
zinc finger protein 142 |
| chr6_+_135513650 | 0.10 |
ENSRNOT00000010676
|
Rcor1
|
REST corepressor 1 |
| chr9_-_100479868 | 0.10 |
ENSRNOT00000022748
|
Pask
|
PAS domain containing serine/threonine kinase |
| chr7_+_25919867 | 0.10 |
ENSRNOT00000009625
ENSRNOT00000090153 |
Ric8b
|
RIC8 guanine nucleotide exchange factor B |
| chr8_-_22974321 | 0.10 |
ENSRNOT00000017369
|
Epor
|
erythropoietin receptor |
| chr6_+_102215603 | 0.10 |
ENSRNOT00000014257
|
Plekhh1
|
pleckstrin homology, MyTH4 and FERM domain containing H1 |
| chrX_-_124870329 | 0.10 |
ENSRNOT00000065023
|
Cul4b
|
cullin 4B |
| chr1_+_267618248 | 0.10 |
ENSRNOT00000017186
|
Gsto2
|
glutathione S-transferase omega 2 |
| chr20_-_12835044 | 0.10 |
ENSRNOT00000074268
|
LOC108348157
|
speriolin-like protein |
| chrX_+_86126157 | 0.10 |
ENSRNOT00000006992
|
Klhl4
|
kelch-like family member 4 |
| chr1_+_211582077 | 0.10 |
ENSRNOT00000023619
|
Pwwp2b
|
PWWP domain containing 2B |
| chr6_-_104631355 | 0.10 |
ENSRNOT00000007825
|
Slc10a1
|
solute carrier family 10 member 1 |
| chr17_+_5311274 | 0.10 |
ENSRNOT00000067020
|
LOC102547665
|
spermatogenesis-associated protein 31D1-like |
| chr14_-_66771750 | 0.10 |
ENSRNOT00000005587
|
Pacrgl
|
PARK2 co-regulated-like |
| chrX_-_14910727 | 0.10 |
ENSRNOT00000085079
ENSRNOT00000030146 |
Ssx1
|
SSX family member 1 |
| chr3_+_154786215 | 0.10 |
ENSRNOT00000019787
|
Lbp
|
lipopolysaccharide binding protein |
| chr5_-_166133491 | 0.10 |
ENSRNOT00000087739
ENSRNOT00000089099 |
Kif1b
|
kinesin family member 1B |
| chr9_+_20765296 | 0.10 |
ENSRNOT00000016291
|
Cd2ap
|
CD2-associated protein |
| chr3_-_80012750 | 0.10 |
ENSRNOT00000018154
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr14_+_39368530 | 0.10 |
ENSRNOT00000084367
|
Cox7b2
|
cytochrome c oxidase subunit 7B2 |
| chr8_+_73682887 | 0.10 |
ENSRNOT00000057522
|
Vps13c
|
vacuolar protein sorting 13C |
| chr20_+_13940877 | 0.10 |
ENSRNOT00000093587
|
Cabin1
|
calcineurin binding protein 1 |
| chr5_-_140024176 | 0.10 |
ENSRNOT00000016492
|
Tmco2
|
transmembrane and coiled-coil domains 2 |
| chr3_+_94530586 | 0.10 |
ENSRNOT00000067860
|
Cstf3
|
cleavage stimulation factor subunit 3 |
| chr8_+_12355767 | 0.10 |
ENSRNOT00000068445
|
Fam76b
|
family with sequence similarity 76, member B |
| chr7_+_114724610 | 0.09 |
ENSRNOT00000014541
|
Dennd3
|
DENN domain containing 3 |
| chr1_+_87563975 | 0.09 |
ENSRNOT00000088772
|
Zfp30
|
zinc finger protein 30 |
| chr10_+_86303727 | 0.09 |
ENSRNOT00000037752
|
Ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr8_-_71728654 | 0.09 |
ENSRNOT00000031207
ENSRNOT00000091751 |
Snx22
|
sorting nexin 22 |
| chr10_+_11912543 | 0.09 |
ENSRNOT00000045192
|
Zfp597
|
zinc finger protein 597 |
| chr2_-_40386669 | 0.09 |
ENSRNOT00000014074
|
Elovl7
|
ELOVL fatty acid elongase 7 |
| chr7_-_15073052 | 0.09 |
ENSRNOT00000037708
|
Zfp799
|
zinc finger protein 799 |
| chr8_+_132032944 | 0.09 |
ENSRNOT00000089278
|
Kif15
|
kinesin family member 15 |
| chr8_-_104995725 | 0.09 |
ENSRNOT00000037120
|
Slc25a36
|
solute carrier family 25 member 36 |
| chr19_-_23554594 | 0.09 |
ENSRNOT00000004590
|
Il15
|
interleukin 15 |
| chr6_-_124735741 | 0.09 |
ENSRNOT00000064716
ENSRNOT00000091693 |
Rps6ka5
|
ribosomal protein S6 kinase A5 |
| chr1_-_199336451 | 0.09 |
ENSRNOT00000035672
|
Prss53
|
protease, serine, 53 |
| chr10_-_63952726 | 0.09 |
ENSRNOT00000090461
|
Doc2b
|
double C2 domain beta |
| chr2_+_207930796 | 0.09 |
ENSRNOT00000047827
|
Kcnd3
|
potassium voltage-gated channel subfamily D member 3 |
| chr9_+_111279495 | 0.09 |
ENSRNOT00000072791
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr7_+_140758615 | 0.09 |
ENSRNOT00000089448
|
Troap
|
trophinin associated protein |
| chr5_+_6373583 | 0.09 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr6_-_102472926 | 0.09 |
ENSRNOT00000079351
|
Zfyve26
|
zinc finger FYVE-type containing 26 |
| chr15_+_24141651 | 0.09 |
ENSRNOT00000082304
|
Lgals3
|
galectin 3 |
| chr10_-_82252720 | 0.09 |
ENSRNOT00000066132
ENSRNOT00000075795 |
Mycbpap
|
Mycbp associated protein |
| chr1_+_211543751 | 0.09 |
ENSRNOT00000049631
|
Lrrc27
|
leucine rich repeat containing 27 |
| chr1_+_99749936 | 0.09 |
ENSRNOT00000025299
|
Klk7
|
kallikrein-related peptidase 7 |
| chr1_-_48565711 | 0.09 |
ENSRNOT00000023387
|
AABR07001512.1
|
|
| chr3_-_44086006 | 0.09 |
ENSRNOT00000034449
ENSRNOT00000082604 |
Ermn
|
ermin |
| chr10_-_82252963 | 0.09 |
ENSRNOT00000086261
|
Mycbpap
|
Mycbp associated protein |
| chr5_+_60900450 | 0.09 |
ENSRNOT00000016384
|
Dcaf10
|
DDB1 and CUL4 associated factor 10 |
| chr10_-_73629581 | 0.09 |
ENSRNOT00000091172
|
Brip1
|
BRCA1 interacting protein C-terminal helicase 1 |
| chr7_-_120882392 | 0.09 |
ENSRNOT00000056179
ENSRNOT00000056178 |
Fam227a
|
family with sequence similarity 227, member A |
| chr5_+_155935554 | 0.09 |
ENSRNOT00000031855
|
LOC100911993
|
ubiquitin carboxyl-terminal hydrolase 48-like |
| chr1_-_62316450 | 0.09 |
ENSRNOT00000079171
|
AABR07001926.3
|
|
| chr3_-_119330345 | 0.09 |
ENSRNOT00000077398
ENSRNOT00000079191 |
Trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr7_+_63922879 | 0.09 |
ENSRNOT00000043581
|
RGD1565498
|
similar to Hypothetical protein LOC270802 |
| chr13_+_70379346 | 0.09 |
ENSRNOT00000038183
|
Nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr1_-_38586563 | 0.09 |
ENSRNOT00000070999
ENSRNOT00000071758 |
LOC501406
|
similar to regulator of sex-limitation candidate 1 |
| chr17_-_43614844 | 0.09 |
ENSRNOT00000023054
|
Hist1h1a
|
histone cluster 1 H1 family member a |
| chr2_-_34313094 | 0.09 |
ENSRNOT00000016863
|
Ppwd1
|
peptidylprolyl isomerase domain and WD repeat containing 1 |
| chr1_+_89220083 | 0.09 |
ENSRNOT00000093144
|
Dmkn
|
dermokine |
| chr17_-_1610745 | 0.09 |
ENSRNOT00000025850
|
Hsd17b3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr20_-_26852199 | 0.09 |
ENSRNOT00000078739
|
Sirt1
|
sirtuin 1 |
| chr1_-_82344345 | 0.09 |
ENSRNOT00000051892
ENSRNOT00000090629 ENSRNOT00000029487 ENSRNOT00000027933 |
Ceacam1
|
carcinoembryonic antigen related cell adhesion molecule 1 |
| chr18_+_57516347 | 0.09 |
ENSRNOT00000082215
|
Gm9949
|
predicted gene 9949 |
| chr12_-_52658275 | 0.09 |
ENSRNOT00000041981
|
Zfp605
|
zinc finger protein 605 |
| chr5_+_78222504 | 0.09 |
ENSRNOT00000019544
|
Slc31a1
|
solute carrier family 31 member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Atf3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1903769 | negative regulation of cell proliferation in bone marrow(GO:1903769) |
| 0.1 | 0.2 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.1 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 0.3 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.1 | 0.2 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.1 | 0.2 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 0.3 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.1 | 0.2 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.1 | 0.2 | GO:1904612 | response to 2,3,7,8-tetrachlorodibenzodioxine(GO:1904612) |
| 0.1 | 0.2 | GO:0000962 | mitochondrial mRNA catabolic process(GO:0000958) positive regulation of mitochondrial RNA catabolic process(GO:0000962) |
| 0.0 | 0.2 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.1 | GO:1904638 | response to resveratrol(GO:1904638) |
| 0.0 | 0.3 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.1 | GO:0045959 | negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 0.2 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.1 | GO:1904412 | regulation of cardiac ventricle development(GO:1904412) |
| 0.0 | 0.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0051256 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:1990792 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 0.0 | 0.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.1 | GO:0061055 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) myotome development(GO:0061055) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.1 | GO:0017143 | insecticide metabolic process(GO:0017143) |
| 0.0 | 0.1 | GO:1904373 | response to kainic acid(GO:1904373) |
| 0.0 | 0.1 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) lipopolysaccharide transport(GO:0015920) |
| 0.0 | 0.1 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.0 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.1 | GO:1904100 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.1 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0071283 | cellular response to iron(III) ion(GO:0071283) |
| 0.0 | 0.1 | GO:0002859 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) cytotoxic T cell degranulation(GO:0043316) positive regulation of activation-induced cell death of T cells(GO:0070237) |
| 0.0 | 0.1 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:1904959 | elastin biosynthetic process(GO:0051542) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.0 | 0.1 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.0 | 0.1 | GO:0015827 | tryptophan transport(GO:0015827) |
| 0.0 | 0.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0032632 | interleukin-3 production(GO:0032632) cadmium ion homeostasis(GO:0055073) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:1902211 | regulation of prolactin signaling pathway(GO:1902211) |
| 0.0 | 0.1 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.0 | 0.1 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.1 | GO:0003275 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.1 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.1 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.0 | 0.1 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.1 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.1 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.1 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.0 | GO:1905077 | regulation of interleukin-17 secretion(GO:1905076) negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.0 | 0.0 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.1 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.0 | 0.1 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.0 | 0.1 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.2 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.1 | GO:0048296 | regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0098705 | copper ion import across plasma membrane(GO:0098705) copper ion import into cell(GO:1902861) |
| 0.0 | 0.1 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.1 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.0 | 0.1 | GO:0060383 | positive regulation of DNA strand elongation(GO:0060383) |
| 0.0 | 0.1 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.0 | 0.1 | GO:1904760 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.2 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.1 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.0 | 0.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.3 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.1 | GO:0015820 | leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.0 | 0.1 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.0 | GO:0043132 | NAD transport(GO:0043132) |
| 0.0 | 0.1 | GO:0060364 | frontal suture morphogenesis(GO:0060364) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) microtubule cytoskeleton organization involved in homologous chromosome segregation(GO:0090172) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.1 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.1 | GO:1904923 | regulation of mitophagy in response to mitochondrial depolarization(GO:1904923) |
| 0.0 | 0.1 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.1 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.0 | 0.1 | GO:0042245 | RNA repair(GO:0042245) |
| 0.0 | 0.1 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.0 | 0.1 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:1905217 | response to astaxanthin(GO:1905217) cellular response to astaxanthin(GO:1905218) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.1 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 0.0 | 0.1 | GO:1904823 | purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:1903173 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.0 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.0 | 0.0 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.0 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.0 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.1 | GO:0021615 | vagus nerve development(GO:0021564) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.0 | 0.0 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.0 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.1 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.0 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.0 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.0 | 0.0 | GO:0002397 | MHC class I protein complex assembly(GO:0002397) |
| 0.0 | 0.1 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.0 | 0.0 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.0 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.0 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 0.0 | 0.1 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.1 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.0 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.0 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.0 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.0 | GO:0035600 | tRNA methylthiolation(GO:0035600) |
| 0.0 | 0.0 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.0 | GO:0021699 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:0071352 | cellular response to interleukin-2(GO:0071352) |
| 0.0 | 0.0 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.0 | GO:2000468 | regulation of peroxidase activity(GO:2000468) |
| 0.0 | 0.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.0 | 0.0 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.1 | GO:0046133 | pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 0.0 | 0.1 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 0.0 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.0 | 0.0 | GO:1902963 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.0 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.0 | 0.1 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 0.0 | 0.0 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.0 | GO:0051595 | response to methylglyoxal(GO:0051595) |
| 0.0 | 0.0 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.0 | 0.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.0 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.0 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.0 | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.1 | GO:2000232 | regulation of rRNA processing(GO:2000232) |
| 0.0 | 0.0 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.0 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.0 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.0 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.0 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.0 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 0.0 | 0.1 | GO:0010044 | response to aluminum ion(GO:0010044) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.0 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.0 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.0 | GO:0070946 | neutrophil mediated killing of gram-positive bacterium(GO:0070946) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.0 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.0 | 0.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0061525 | hindgut development(GO:0061525) |
| 0.0 | 0.0 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.1 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0045025 | mitochondrial degradosome(GO:0045025) |
| 0.0 | 0.1 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.2 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.2 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.1 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.5 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.0 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.1 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0090534 | calcium ion-transporting ATPase complex(GO:0090534) |
| 0.0 | 0.0 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.1 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0071008 | B cell receptor complex(GO:0019815) U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 0.0 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.0 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.0 | GO:0070110 | interleukin-6 receptor complex(GO:0005896) ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.0 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.0 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.0 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.1 | 0.2 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.1 | 0.1 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.2 | GO:0019863 | IgE binding(GO:0019863) advanced glycation end-product receptor activity(GO:0050785) |
| 0.0 | 0.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0004956 | prostaglandin D receptor activity(GO:0004956) |
| 0.0 | 0.1 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.0 | 0.3 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.2 | GO:1901612 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.1 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 0.1 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.1 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) |
| 0.0 | 0.1 | GO:0045174 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.1 | GO:0032767 | copper-exporting ATPase activity(GO:0004008) copper-dependent protein binding(GO:0032767) copper-transporting ATPase activity(GO:0043682) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.2 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.1 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.0 | 0.1 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.0 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.0 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.0 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.0 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.1 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.0 | 0.0 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0016726 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.0 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.0 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0005111 | type 1 fibroblast growth factor receptor binding(GO:0005105) type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.0 | GO:0035596 | methylthiotransferase activity(GO:0035596) transferase activity, transferring alkylthio groups(GO:0050497) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.0 | GO:0001565 | phorbol ester receptor activity(GO:0001565) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.0 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.0 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.0 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.0 | 0.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.0 | GO:0102390 | mycophenolic acid acyl-glucuronide esterase activity(GO:0102390) |
| 0.0 | 0.0 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.0 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.1 | GO:0043734 | DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.0 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.0 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.0 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.0 | GO:0000827 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.0 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.5 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.3 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.1 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.0 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |