Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
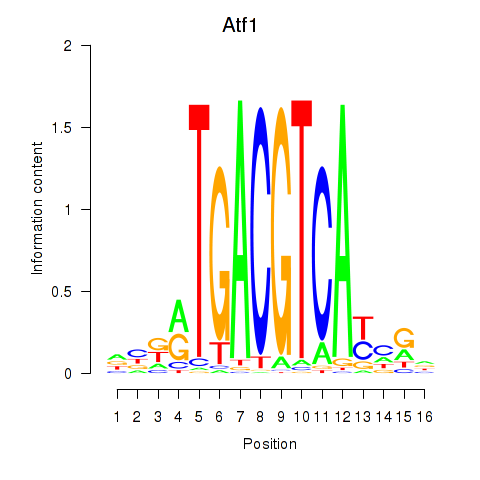
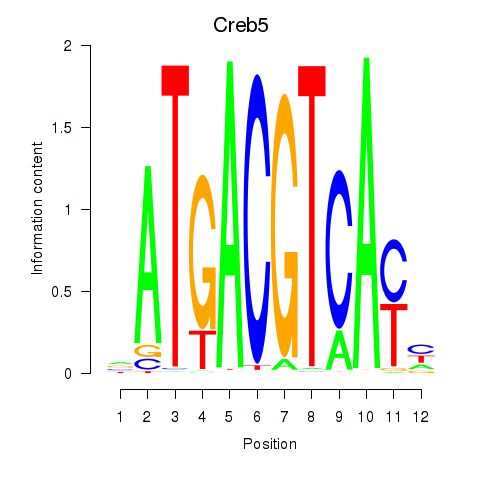
Results for Atf1_Creb5
Z-value: 0.59
Transcription factors associated with Atf1_Creb5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Atf1
|
ENSRNOG00000061088 | activating transcription factor 1 |
|
Creb5
|
ENSRNOG00000008622 | cAMP responsive element binding protein 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Creb5 | rn6_v1_chr4_+_83713666_83713666 | 0.76 | 1.4e-01 | Click! |
| Atf1 | rn6_v1_chr7_+_141882251_141882251 | -0.70 | 1.9e-01 | Click! |
Activity profile of Atf1_Creb5 motif
Sorted Z-values of Atf1_Creb5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_150801289 | 0.64 |
ENSRNOT00000035060
|
Map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr7_+_120580743 | 0.51 |
ENSRNOT00000017181
|
Maff
|
MAF bZIP transcription factor F |
| chr10_+_70884531 | 0.49 |
ENSRNOT00000015199
|
Ccl4
|
C-C motif chemokine ligand 4 |
| chr20_-_45815940 | 0.37 |
ENSRNOT00000073276
|
Gpr6
|
G protein-coupled receptor 6 |
| chr10_+_16970626 | 0.36 |
ENSRNOT00000005383
|
Dusp1
|
dual specificity phosphatase 1 |
| chr2_+_164549455 | 0.36 |
ENSRNOT00000017151
|
Mlf1
|
myeloid leukemia factor 1 |
| chr7_-_44121130 | 0.33 |
ENSRNOT00000005706
|
Nts
|
neurotensin |
| chr9_+_10535340 | 0.25 |
ENSRNOT00000075408
|
Znrf4
|
zinc and ring finger 4 |
| chr16_+_72401887 | 0.25 |
ENSRNOT00000074449
|
LOC100910163
|
uncharacterized LOC100910163 |
| chr9_-_113358526 | 0.24 |
ENSRNOT00000065073
|
Txndc2
|
thioredoxin domain containing 2 |
| chr17_-_9695292 | 0.24 |
ENSRNOT00000036162
|
Prr7
|
proline rich 7 (synaptic) |
| chr12_+_2170630 | 0.24 |
ENSRNOT00000071928
|
Pet100
|
PET100 homolog |
| chr6_-_91456696 | 0.23 |
ENSRNOT00000005577
|
Rps29
|
ribosomal protein S29 |
| chr1_-_80221710 | 0.23 |
ENSRNOT00000091687
|
Fosb
|
FosB proto-oncogene, AP-1 transcription factor subunit |
| chr1_+_211582077 | 0.23 |
ENSRNOT00000023619
|
Pwwp2b
|
PWWP domain containing 2B |
| chr8_+_64364741 | 0.22 |
ENSRNOT00000082840
|
Celf6
|
CUGBP, Elav-like family member 6 |
| chr16_-_49574314 | 0.22 |
ENSRNOT00000017568
ENSRNOT00000085535 ENSRNOT00000017054 |
Pdlim3
|
PDZ and LIM domain 3 |
| chr10_+_89376530 | 0.22 |
ENSRNOT00000028089
|
Rnd2
|
Rho family GTPase 2 |
| chr6_+_1657331 | 0.21 |
ENSRNOT00000049672
ENSRNOT00000079864 |
Qpct
|
glutaminyl-peptide cyclotransferase |
| chr11_+_88424414 | 0.21 |
ENSRNOT00000022328
|
Spag6l
|
sperm associated antigen 6-like |
| chrX_+_114929029 | 0.20 |
ENSRNOT00000006459
|
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr4_+_83391283 | 0.20 |
ENSRNOT00000031365
|
Creb5
|
cAMP responsive element binding protein 5 |
| chr6_-_47848026 | 0.20 |
ENSRNOT00000011048
ENSRNOT00000090017 |
Allc
|
allantoicase |
| chr1_-_89190128 | 0.19 |
ENSRNOT00000067813
|
Gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr8_+_58431407 | 0.19 |
ENSRNOT00000011974
|
Sln
|
sarcolipin |
| chr2_-_251970768 | 0.19 |
ENSRNOT00000020141
|
Wdr63
|
WD repeat domain 63 |
| chr16_+_83161880 | 0.18 |
ENSRNOT00000080793
|
1700016D06Rik
|
RIKEN cDNA 1700016D06 gene |
| chr4_+_67378188 | 0.17 |
ENSRNOT00000030892
|
Ndufb2
|
NADH:ubiquinone oxidoreductase subunit B2 |
| chr1_-_220467159 | 0.17 |
ENSRNOT00000075365
|
Tmem151a
|
transmembrane protein 151A |
| chr1_-_101118825 | 0.17 |
ENSRNOT00000066328
|
Rps11
|
ribosomal protein S11 |
| chr1_-_89179825 | 0.17 |
ENSRNOT00000028514
|
Tmem147
|
transmembrane protein 147 |
| chr3_+_151285249 | 0.16 |
ENSRNOT00000055254
|
Procr
|
protein C receptor |
| chr5_-_107857320 | 0.16 |
ENSRNOT00000008898
|
Cdkn2b
|
cyclin-dependent kinase inhibitor 2B |
| chr10_-_14056169 | 0.16 |
ENSRNOT00000017833
|
Syngr3
|
synaptogyrin 3 |
| chr5_+_159845774 | 0.16 |
ENSRNOT00000012328
|
Epha2
|
Eph receptor A2 |
| chr10_+_55924938 | 0.15 |
ENSRNOT00000087003
ENSRNOT00000057079 |
Trappc1
|
trafficking protein particle complex 1 |
| chr8_+_44136496 | 0.15 |
ENSRNOT00000087022
|
Scn3b
|
sodium voltage-gated channel beta subunit 3 |
| chr12_-_2007516 | 0.15 |
ENSRNOT00000037564
|
Pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr17_+_10537365 | 0.15 |
ENSRNOT00000023651
|
Cltb
|
clathrin, light chain B |
| chr15_+_28319136 | 0.15 |
ENSRNOT00000048723
|
Tppp2
|
tubulin polymerization-promoting protein family member 2 |
| chr5_-_146446227 | 0.14 |
ENSRNOT00000044868
|
Hmgb4
|
high-mobility group box 4 |
| chr15_+_52265557 | 0.14 |
ENSRNOT00000015969
|
Nudt18
|
nudix hydrolase 18 |
| chr19_+_54245950 | 0.14 |
ENSRNOT00000024033
|
Cox4i1
|
cytochrome c oxidase subunit 4i1 |
| chr15_-_70399924 | 0.14 |
ENSRNOT00000087940
|
Diaph3
|
diaphanous-related formin 3 |
| chr6_+_8669722 | 0.14 |
ENSRNOT00000048550
|
Camkmt
|
calmodulin-lysine N-methyltransferase |
| chr1_+_211205903 | 0.14 |
ENSRNOT00000023139
|
Ppp2r2d
|
protein phosphatase 2, regulatory subunit B, delta |
| chr9_-_85243001 | 0.13 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr1_+_201429771 | 0.13 |
ENSRNOT00000027836
|
Plekha1
|
pleckstrin homology domain containing A1 |
| chr5_-_148577292 | 0.13 |
ENSRNOT00000017156
|
Zcchc17
|
zinc finger CCHC-type containing 17 |
| chr9_+_17817721 | 0.13 |
ENSRNOT00000086986
ENSRNOT00000026920 |
Hsp90ab1
|
heat shock protein 90 alpha family class B member 1 |
| chr10_-_13542077 | 0.12 |
ENSRNOT00000008736
|
Atp6v0c
|
ATPase H+ transporting V0 subunit C |
| chr11_+_84094520 | 0.12 |
ENSRNOT00000046642
|
Rps15al2
|
ribosomal protein S15A-like 2 |
| chr10_+_56576428 | 0.12 |
ENSRNOT00000079237
ENSRNOT00000023291 |
Cldn7
|
claudin 7 |
| chr13_-_50916982 | 0.12 |
ENSRNOT00000004408
|
Btg2
|
BTG anti-proliferation factor 2 |
| chr13_-_110257367 | 0.12 |
ENSRNOT00000005576
|
Dtl
|
denticleless E3 ubiquitin protein ligase homolog |
| chr1_-_89194602 | 0.12 |
ENSRNOT00000028518
|
Gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr19_-_54245855 | 0.12 |
ENSRNOT00000023855
|
Emc8
|
ER membrane protein complex subunit 8 |
| chr2_-_187909394 | 0.12 |
ENSRNOT00000032355
|
Rab25
|
RAB25, member RAS oncogene family |
| chr11_+_86890585 | 0.12 |
ENSRNOT00000002579
|
Ranbp1
|
RAN binding protein 1 |
| chr3_-_161212188 | 0.12 |
ENSRNOT00000065751
|
Wfdc3
|
WAP four-disulfide core domain 3 |
| chrX_-_139329975 | 0.12 |
ENSRNOT00000086405
|
LOC103694517
|
high mobility group protein B4-like |
| chr6_+_86131242 | 0.11 |
ENSRNOT00000039337
|
Fau
|
FAU, ubiquitin like and ribosomal protein S30 fusion |
| chr14_-_18531179 | 0.11 |
ENSRNOT00000003703
|
Areg
|
amphiregulin |
| chr10_-_59892960 | 0.11 |
ENSRNOT00000084432
|
Aspa
|
aspartoacylase |
| chr4_+_40161285 | 0.11 |
ENSRNOT00000050722
|
LOC500035
|
hypothetical protein LOC500035 |
| chr6_+_98284170 | 0.11 |
ENSRNOT00000031979
|
Rhoj
|
ras homolog family member J |
| chr1_+_73719005 | 0.11 |
ENSRNOT00000025110
|
Cdc42ep5
|
CDC42 effector protein 5 |
| chr8_+_22021213 | 0.11 |
ENSRNOT00000049706
|
Mrpl4
|
mitochondrial ribosomal protein L4 |
| chr9_+_102862890 | 0.10 |
ENSRNOT00000050494
ENSRNOT00000080129 |
Fam174a
|
family with sequence similarity 174, member A |
| chr14_+_10581136 | 0.10 |
ENSRNOT00000002987
|
Coq2
|
coenzyme Q2, polyprenyltransferase |
| chr10_-_64398294 | 0.10 |
ENSRNOT00000010386
|
Glod4
|
glyoxalase domain containing 4 |
| chr1_+_102900286 | 0.10 |
ENSRNOT00000017468
|
Ldha
|
lactate dehydrogenase A |
| chr4_+_69386698 | 0.10 |
ENSRNOT00000091655
|
Trbv13-2
|
T cell receptor beta, variable 13-2 |
| chr10_-_88163712 | 0.10 |
ENSRNOT00000005382
ENSRNOT00000084493 |
Krt17
|
keratin 17 |
| chr13_-_102857551 | 0.10 |
ENSRNOT00000080309
|
Mark1
|
microtubule affinity regulating kinase 1 |
| chr4_+_70977556 | 0.10 |
ENSRNOT00000031984
|
LOC680112
|
hypothetical protein LOC680112 |
| chr3_-_45210474 | 0.10 |
ENSRNOT00000091777
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr7_-_70355619 | 0.10 |
ENSRNOT00000031272
|
Tspan31
|
tetraspanin 31 |
| chr20_-_10680283 | 0.10 |
ENSRNOT00000001579
|
Sik1
|
salt-inducible kinase 1 |
| chr19_-_15733412 | 0.10 |
ENSRNOT00000014831
|
Irx6
|
iroquois homeobox 6 |
| chr12_+_47024442 | 0.10 |
ENSRNOT00000001545
|
Cox6a1
|
cytochrome c oxidase subunit 6A1 |
| chr8_-_87282156 | 0.10 |
ENSRNOT00000087874
|
Filip1
|
filamin A interacting protein 1 |
| chr1_-_198267093 | 0.10 |
ENSRNOT00000047477
|
RGD1563217
|
similar to RIKEN cDNA 4930451I11 |
| chr5_-_151397603 | 0.10 |
ENSRNOT00000076866
|
Gpr3
|
G protein-coupled receptor 3 |
| chr9_-_78969013 | 0.10 |
ENSRNOT00000019772
ENSRNOT00000057585 |
Fn1
|
fibronectin 1 |
| chr17_+_57075218 | 0.10 |
ENSRNOT00000089536
|
Crem
|
cAMP responsive element modulator |
| chr1_+_280423079 | 0.10 |
ENSRNOT00000011983
|
Slc18a2
|
solute carrier family 18 member A2 |
| chr16_+_20317446 | 0.10 |
ENSRNOT00000025626
|
Ccdc124
|
coiled-coil domain containing 124 |
| chr10_+_37594824 | 0.09 |
ENSRNOT00000085463
|
Skp1
|
S-phase kinase-associated protein 1 |
| chr5_-_141430659 | 0.09 |
ENSRNOT00000034944
|
Akirin1
|
akirin 1 |
| chr10_+_10967658 | 0.09 |
ENSRNOT00000004999
|
Cdip1
|
cell death-inducing p53 target 1 |
| chr7_+_10962330 | 0.09 |
ENSRNOT00000008360
|
Tle6
|
transducin-like enhancer of split 6 |
| chr15_-_43733182 | 0.09 |
ENSRNOT00000015318
|
Ppp2r2a
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr2_-_123281856 | 0.09 |
ENSRNOT00000079745
|
Ccna2
|
cyclin A2 |
| chr10_-_36716601 | 0.09 |
ENSRNOT00000038838
|
LOC497899
|
similar to hypothetical protein 4930503F14 |
| chr1_-_103323476 | 0.09 |
ENSRNOT00000019051
|
Mrgprx3
|
MAS related GPR family member X3 |
| chr17_+_57074525 | 0.09 |
ENSRNOT00000020012
ENSRNOT00000074146 |
Crem
|
cAMP responsive element modulator |
| chr20_-_3299580 | 0.09 |
ENSRNOT00000050373
|
Gnl1
|
G protein nucleolar 1 |
| chr1_+_142087208 | 0.09 |
ENSRNOT00000017532
|
Prc1
|
protein regulator of cytokinesis 1 |
| chr10_-_65424802 | 0.09 |
ENSRNOT00000018468
|
Traf4
|
Tnf receptor associated factor 4 |
| chr19_+_16415636 | 0.09 |
ENSRNOT00000089975
|
Irx5
|
iroquois homeobox 5 |
| chr1_+_213577122 | 0.09 |
ENSRNOT00000071925
|
RGD1309350
|
similar to transthyretin (4L369) |
| chr13_-_83504494 | 0.09 |
ENSRNOT00000004083
|
Tiprl
|
TOR signaling pathway regulator |
| chr10_+_102136283 | 0.09 |
ENSRNOT00000003735
|
Sstr2
|
somatostatin receptor 2 |
| chr1_+_167309051 | 0.09 |
ENSRNOT00000055235
ENSRNOT00000076894 |
Pgap2
|
post-GPI attachment to proteins 2 |
| chr5_-_25584278 | 0.08 |
ENSRNOT00000090579
ENSRNOT00000090376 |
Pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr4_+_168832910 | 0.08 |
ENSRNOT00000011134
|
Gprc5a
|
G protein-coupled receptor, class C, group 5, member A |
| chr10_+_37594578 | 0.08 |
ENSRNOT00000007676
|
Skp1
|
S-phase kinase-associated protein 1 |
| chr9_-_99651813 | 0.08 |
ENSRNOT00000022089
|
Ndufa10
|
NADH:ubiquinone oxidoreductase subunit A10 |
| chr2_+_181331464 | 0.08 |
ENSRNOT00000017448
|
Map9
|
microtubule-associated protein 9 |
| chr2_+_188784222 | 0.08 |
ENSRNOT00000028095
|
Pmvk
|
phosphomevalonate kinase |
| chr19_-_25955371 | 0.08 |
ENSRNOT00000004042
ENSRNOT00000084123 |
Rad23a
|
RAD23 homolog A, nucleotide excision repair protein |
| chr4_-_50312608 | 0.08 |
ENSRNOT00000010019
|
Fezf1
|
Fez family zinc finger 1 |
| chr3_+_152933771 | 0.08 |
ENSRNOT00000027539
|
RGD1307752
|
similar to RIKEN cDNA 1110008F13 |
| chrX_-_132424746 | 0.08 |
ENSRNOT00000087819
|
LOC681067
|
similar to RIKEN cDNA 1700001F22 |
| chrX_-_158261717 | 0.08 |
ENSRNOT00000086804
|
RGD1561558
|
similar to RIKEN cDNA 1700001F22 |
| chr2_+_86951776 | 0.08 |
ENSRNOT00000087275
|
AABR07009105.1
|
|
| chrX_+_28593405 | 0.08 |
ENSRNOT00000071708
|
Tmsb4x
|
thymosin beta 4, X-linked |
| chr2_-_66608324 | 0.08 |
ENSRNOT00000077597
|
Cln5
|
ceroid-lipofuscinosis, neuronal 5 |
| chr2_+_208738132 | 0.08 |
ENSRNOT00000023972
|
AABR07012795.1
|
|
| chr1_-_89303968 | 0.08 |
ENSRNOT00000056714
|
Ffar3
|
free fatty acid receptor 3 |
| chr7_-_2909144 | 0.08 |
ENSRNOT00000082518
ENSRNOT00000089074 ENSRNOT00000085644 |
Myl6
|
myosin light chain 6 |
| chr17_+_44763598 | 0.08 |
ENSRNOT00000079880
|
Hist1h3b
|
histone cluster 1, H3b |
| chr1_-_80221417 | 0.08 |
ENSRNOT00000072149
|
Fosb
|
FosB proto-oncogene, AP-1 transcription factor subunit |
| chr4_-_120559078 | 0.08 |
ENSRNOT00000085730
ENSRNOT00000079575 |
Kbtbd12
|
kelch repeat and BTB domain containing 12 |
| chr13_+_53283855 | 0.08 |
ENSRNOT00000043590
|
RGD1564839
|
similar to ribosomal protein L31 |
| chr1_-_101514547 | 0.08 |
ENSRNOT00000079633
|
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr1_-_47213749 | 0.08 |
ENSRNOT00000024656
|
Dynlt1
|
dynein light chain Tctex-type 1 |
| chrX_-_144001727 | 0.07 |
ENSRNOT00000078404
|
RGD1560585
|
similar to RIKEN cDNA 1700001F22 |
| chr16_-_82439441 | 0.07 |
ENSRNOT00000040315
|
AABR07026539.1
|
|
| chr13_+_52553775 | 0.07 |
ENSRNOT00000011991
|
Csrp1
|
cysteine and glycine-rich protein 1 |
| chr8_-_21995806 | 0.07 |
ENSRNOT00000028034
|
S1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr1_+_100473643 | 0.07 |
ENSRNOT00000026379
|
Josd2
|
Josephin domain containing 2 |
| chr1_+_221420271 | 0.07 |
ENSRNOT00000028481
|
LOC687780
|
similar to Finkel-Biskis-Reilly murine sarcoma virusubiquitously expressed |
| chr17_+_31493107 | 0.07 |
ENSRNOT00000023611
ENSRNOT00000086264 |
Tubb2a
|
tubulin, beta 2A class IIa |
| chr20_-_10844178 | 0.07 |
ENSRNOT00000079207
|
Hsf2bp
|
heat shock transcription factor 2 binding protein |
| chr4_-_170740274 | 0.07 |
ENSRNOT00000012212
|
Gucy2c
|
guanylate cyclase 2C |
| chr3_-_120011364 | 0.07 |
ENSRNOT00000018922
|
Fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr12_-_6879154 | 0.07 |
ENSRNOT00000001207
|
Alox5ap
|
arachidonate 5-lipoxygenase activating protein |
| chrX_+_106774980 | 0.07 |
ENSRNOT00000046091
|
Tceal7
|
transcription elongation factor A like 7 |
| chr1_-_64021321 | 0.07 |
ENSRNOT00000090819
|
Rps9
|
ribosomal protein S9 |
| chr19_-_43841795 | 0.07 |
ENSRNOT00000079539
|
Ldhd
|
lactate dehydrogenase D |
| chr1_+_100755682 | 0.07 |
ENSRNOT00000035748
|
Vrk3
|
vaccinia related kinase 3 |
| chr6_-_135049728 | 0.07 |
ENSRNOT00000009556
|
Hsp90aa1
|
heat shock protein 90 alpha family class A member 1 |
| chr16_-_20873344 | 0.07 |
ENSRNOT00000027381
|
Cope
|
coatomer protein complex, subunit epsilon |
| chr1_+_80982358 | 0.07 |
ENSRNOT00000078462
|
AABR07002683.1
|
|
| chr6_-_126622532 | 0.07 |
ENSRNOT00000038816
|
Moap1
|
modulator of apoptosis 1 |
| chr11_-_64583994 | 0.07 |
ENSRNOT00000004289
|
B4galt4
|
beta-1,4-galactosyltransferase 4 |
| chr14_-_86739335 | 0.07 |
ENSRNOT00000078282
|
Purb
|
purine rich element binding protein B |
| chr5_-_152464850 | 0.07 |
ENSRNOT00000021937
|
Zfp593
|
zinc finger protein 593 |
| chr11_-_36479868 | 0.07 |
ENSRNOT00000075762
|
LOC100911295
|
non-histone chromosomal protein HMG-14-like |
| chr19_-_37990353 | 0.07 |
ENSRNOT00000026817
|
Ddx28
|
DEAD-box helicase 28 |
| chr19_+_49457677 | 0.07 |
ENSRNOT00000084445
|
Cenpn
|
centromere protein N |
| chr10_-_48599208 | 0.06 |
ENSRNOT00000003974
|
Zswim7
|
zinc finger, SWIM-type containing 7 |
| chr4_-_183697531 | 0.06 |
ENSRNOT00000055441
|
Amn1
|
antagonist of mitotic exit network 1 homolog |
| chrX_-_82986051 | 0.06 |
ENSRNOT00000077587
|
Hdx
|
highly divergent homeobox |
| chr4_+_157453069 | 0.06 |
ENSRNOT00000088622
|
Mlf2
|
myeloid leukemia factor 2 |
| chr19_+_56272162 | 0.06 |
ENSRNOT00000030399
|
Afg3l1
|
AFG3(ATPase family gene 3)-like 1 (S. cerevisiae) |
| chr14_-_6900733 | 0.06 |
ENSRNOT00000061224
|
Dmp1
|
dentin matrix acidic phosphoprotein 1 |
| chr2_+_233602732 | 0.06 |
ENSRNOT00000044232
|
Pitx2
|
paired-like homeodomain 2 |
| chr1_-_37863756 | 0.06 |
ENSRNOT00000077781
|
Zfp874b
|
zinc finger protein 874b |
| chr9_-_82673898 | 0.06 |
ENSRNOT00000027165
|
Chpf
|
chondroitin polymerizing factor |
| chr13_-_50499060 | 0.06 |
ENSRNOT00000065347
ENSRNOT00000076924 |
Etnk2
|
ethanolamine kinase 2 |
| chr15_+_24141651 | 0.06 |
ENSRNOT00000082304
|
Lgals3
|
galectin 3 |
| chr2_+_187347602 | 0.06 |
ENSRNOT00000025384
|
Nes
|
nestin |
| chr13_-_104284068 | 0.06 |
ENSRNOT00000005407
|
LOC100912365
|
uncharacterized LOC100912365 |
| chr3_+_103945329 | 0.06 |
ENSRNOT00000008364
|
Emc7
|
ER membrane protein complex subunit 7 |
| chr17_+_82066152 | 0.06 |
ENSRNOT00000083034
|
Arl5b
|
ADP-ribosylation factor like GTPase 5B |
| chr19_+_55917736 | 0.06 |
ENSRNOT00000020635
|
Rpl13
|
ribosomal protein L13 |
| chr14_+_35683442 | 0.06 |
ENSRNOT00000003085
|
Chic2
|
cysteine-rich hydrophobic domain 2 |
| chr19_+_27404712 | 0.06 |
ENSRNOT00000023657
|
Mylk3
|
myosin light chain kinase 3 |
| chr15_-_39886613 | 0.06 |
ENSRNOT00000089963
|
Cdadc1
|
cytidine and dCMP deaminase domain containing 1 |
| chr9_+_82370924 | 0.06 |
ENSRNOT00000025219
|
Zfand2b
|
zinc finger AN1-type containing 2B |
| chr2_-_28799266 | 0.06 |
ENSRNOT00000089293
|
Tmem171
|
transmembrane protein 171 |
| chr14_-_86706626 | 0.06 |
ENSRNOT00000082893
|
H2afv
|
H2A histone family, member V |
| chr7_-_124982566 | 0.06 |
ENSRNOT00000075099
|
Sult4a1
|
sulfotransferase family 4A, member 1 |
| chr1_-_67302751 | 0.06 |
ENSRNOT00000041518
|
Vom1r42
|
vomeronasal 1 receptor 42 |
| chr17_+_78915604 | 0.06 |
ENSRNOT00000057855
|
Rpp38
|
ribonuclease P/MRP 38 subunit |
| chr1_+_225048149 | 0.06 |
ENSRNOT00000036454
|
Lrrn4cl
|
LRRN4 C-terminal like |
| chrX_+_16170576 | 0.06 |
ENSRNOT00000003895
|
Clcn5
|
chloride voltage-gated channel 5 |
| chr7_-_14303055 | 0.06 |
ENSRNOT00000008963
|
Brd4
|
bromodomain containing 4 |
| chr16_-_20534209 | 0.06 |
ENSRNOT00000026601
|
Mk1
|
Mk1 protein |
| chr15_+_5904569 | 0.06 |
ENSRNOT00000072599
|
LOC102546376
|
disks large homolog 5-like |
| chr6_-_135112775 | 0.06 |
ENSRNOT00000086310
|
LOC103692716
|
heat shock protein HSP 90-alpha |
| chr10_+_55492404 | 0.06 |
ENSRNOT00000005588
ENSRNOT00000078038 |
Rpl26
|
ribosomal protein L26 |
| chr2_+_104290726 | 0.06 |
ENSRNOT00000017387
|
Dnajc5b
|
DnaJ heat shock protein family (Hsp40) member C5 beta |
| chr15_+_43007908 | 0.06 |
ENSRNOT00000084753
ENSRNOT00000091567 ENSRNOT00000087709 |
Stmn4
|
stathmin 4 |
| chr17_+_57040023 | 0.06 |
ENSRNOT00000020204
|
Crem
|
cAMP responsive element modulator |
| chr4_-_182600258 | 0.05 |
ENSRNOT00000067970
|
Ergic2
|
ERGIC and golgi 2 |
| chr13_+_111890894 | 0.05 |
ENSRNOT00000007341
|
LOC100125367
|
hypothetical protein LOC100125367 |
| chr6_+_26780352 | 0.05 |
ENSRNOT00000009916
|
Prr30
|
proline rich 30 |
| chr2_-_264864265 | 0.05 |
ENSRNOT00000044236
|
Srsf11
|
serine and arginine rich splicing factor 11 |
| chr20_+_2088533 | 0.05 |
ENSRNOT00000001012
ENSRNOT00000079021 |
Znrd1
|
zinc ribbon domain containing 1 |
| chr14_-_37361798 | 0.05 |
ENSRNOT00000002980
|
Cwh43
|
cell wall biogenesis 43 C-terminal homolog |
| chr4_-_80333326 | 0.05 |
ENSRNOT00000014058
|
LOC100363502
|
cytochrome c, somatic-like |
| chr5_-_58163584 | 0.05 |
ENSRNOT00000060594
|
Ccl27
|
C-C motif chemokine ligand 27 |
| chr1_+_209769282 | 0.05 |
ENSRNOT00000083316
|
Glrx3
|
glutaredoxin 3 |
| chr2_-_30748325 | 0.05 |
ENSRNOT00000084294
ENSRNOT00000083089 |
Mrps36
|
mitochondrial ribosomal protein S36 |
| chr3_-_160922341 | 0.05 |
ENSRNOT00000029206
|
Tp53tg5
|
TP53 target 5 |
| chr16_+_90613870 | 0.05 |
ENSRNOT00000079334
|
Shcbp1
|
SHC binding and spindle associated 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Atf1_Creb5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.2 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 0.2 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.1 | 0.3 | GO:0097332 | response to antipsychotic drug(GO:0097332) |
| 0.1 | 0.2 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.1 | 0.3 | GO:0061741 | vacuolar transmembrane transport(GO:0034486) chaperone-mediated protein transport involved in chaperone-mediated autophagy(GO:0061741) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) |
| 0.0 | 0.1 | GO:0072702 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) response to methyl methanesulfonate(GO:0072702) cellular response to methyl methanesulfonate(GO:0072703) response to environmental enrichment(GO:0090648) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.6 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.2 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.1 | GO:0007161 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) calcium-independent cell-matrix adhesion(GO:0007161) interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.0 | 0.4 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0002877 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.0 | 0.4 | GO:0090266 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.2 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.1 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.0 | 0.1 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.0 | 0.1 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.2 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.1 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.1 | GO:0060460 | pulmonary myocardium development(GO:0003350) subthalamus development(GO:0021539) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.1 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.1 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.2 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.1 | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 0.0 | 0.1 | GO:0019660 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.1 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.0 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.1 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.1 | GO:0045105 | intermediate filament polymerization or depolymerization(GO:0045105) |
| 0.0 | 0.1 | GO:0006735 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.2 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.0 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 0.0 | GO:0021629 | muscle attachment(GO:0016203) olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.2 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.1 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.0 | GO:0052422 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.2 | GO:0060253 | negative regulation of glial cell proliferation(GO:0060253) |
| 0.0 | 0.0 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.0 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.0 | 0.0 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.0 | GO:0033505 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.0 | 0.2 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.0 | 0.1 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:1901977 | negative regulation of cell cycle checkpoint(GO:1901977) negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.0 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.0 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.0 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.1 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.6 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.1 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.0 | 0.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.1 | GO:0030681 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.3 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.0 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.4 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.0 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.5 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.3 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.6 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.2 | GO:0002134 | UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 0.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.1 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.1 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.0 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0050785 | IgE binding(GO:0019863) advanced glycation end-product receptor activity(GO:0050785) |
| 0.0 | 0.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.0 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.0 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.0 | GO:0032564 | dATP binding(GO:0032564) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.0 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |