Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
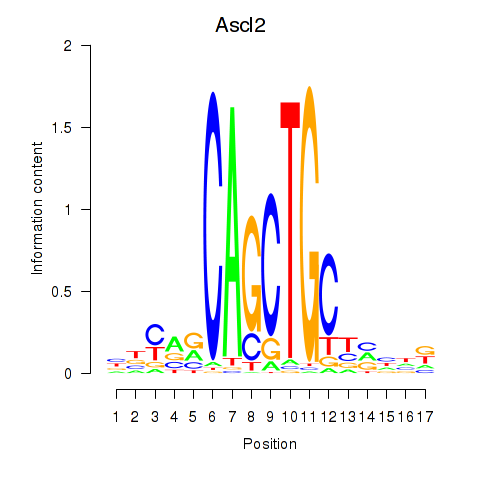
Results for Ascl2
Z-value: 0.82
Transcription factors associated with Ascl2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ascl2
|
ENSRNOG00000020434 | achaete-scute family bHLH transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ascl2 | rn6_v1_chr1_-_216156409_216156409 | 0.72 | 1.7e-01 | Click! |
Activity profile of Ascl2 motif
Sorted Z-values of Ascl2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_21345805 | 0.95 |
ENSRNOT00000081296
ENSRNOT00000007802 |
Car8
|
carbonic anhydrase 8 |
| chr5_-_151459037 | 0.91 |
ENSRNOT00000064472
ENSRNOT00000087836 |
Sytl1
|
synaptotagmin-like 1 |
| chr4_+_86275717 | 0.83 |
ENSRNOT00000016414
|
Ppp1r17
|
protein phosphatase 1, regulatory subunit 17 |
| chr16_+_10250404 | 0.64 |
ENSRNOT00000087037
ENSRNOT00000081183 |
Gdf10
|
growth differentiation factor 10 |
| chr14_+_4362717 | 0.64 |
ENSRNOT00000002887
|
Barhl2
|
BarH-like homeobox 2 |
| chr16_-_20807070 | 0.56 |
ENSRNOT00000072536
|
Comp
|
cartilage oligomeric matrix protein |
| chr1_+_90948976 | 0.44 |
ENSRNOT00000056877
|
LOC108348111
|
succinate dehydrogenase assembly factor 1, mitochondrial |
| chr7_-_3342491 | 0.43 |
ENSRNOT00000081756
|
Rdh5
|
retinol dehydrogenase 5 |
| chr5_-_156734541 | 0.41 |
ENSRNOT00000021036
|
LOC100909857
|
cytidine deaminase-like |
| chr8_-_115167486 | 0.41 |
ENSRNOT00000033018
|
Gpr62
|
G protein-coupled receptor 62 |
| chr7_+_72924799 | 0.34 |
ENSRNOT00000008969
|
Laptm4b
|
lysosomal protein transmembrane 4 beta |
| chr16_-_59366824 | 0.31 |
ENSRNOT00000015062
|
RGD1304810
|
similar to 6430573F11Rik protein |
| chr4_-_150244372 | 0.30 |
ENSRNOT00000047685
|
Ret
|
ret proto-oncogene |
| chr19_-_52282877 | 0.30 |
ENSRNOT00000021271
|
Kcng4
|
potassium voltage-gated channel modifier subfamily G member 4 |
| chr6_+_98284170 | 0.29 |
ENSRNOT00000031979
|
Rhoj
|
ras homolog family member J |
| chr19_-_55183557 | 0.27 |
ENSRNOT00000017317
|
Mlnr
|
motilin receptor |
| chr3_+_2480232 | 0.27 |
ENSRNOT00000014489
|
Tprn
|
taperin |
| chr6_+_58467254 | 0.27 |
ENSRNOT00000065396
|
Etv1
|
ets variant 1 |
| chr1_+_100199057 | 0.27 |
ENSRNOT00000025831
|
Klk1
|
kallikrein 1 |
| chr10_+_109278712 | 0.26 |
ENSRNOT00000065565
|
LOC690871
|
hypothetical protein LOC690871 |
| chr16_-_6669045 | 0.26 |
ENSRNOT00000067639
|
Prkcd
|
protein kinase C, delta |
| chr1_-_277768368 | 0.24 |
ENSRNOT00000023113
|
Afap1l2
|
actin filament associated protein 1-like 2 |
| chr7_-_130120579 | 0.24 |
ENSRNOT00000044376
|
Mapk12
|
mitogen-activated protein kinase 12 |
| chr11_+_17538063 | 0.23 |
ENSRNOT00000031889
ENSRNOT00000090878 |
Chodl
|
chondrolectin |
| chr9_+_10339075 | 0.22 |
ENSRNOT00000073402
|
Vmac
|
vimentin-type intermediate filament associated coiled-coil protein |
| chr5_-_155772040 | 0.22 |
ENSRNOT00000036788
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr1_-_89560719 | 0.22 |
ENSRNOT00000028653
|
Scn1b
|
sodium voltage-gated channel beta subunit 1 |
| chr1_+_221470674 | 0.21 |
ENSRNOT00000054835
|
Naaladl1
|
N-acetylated alpha-linked acidic dipeptidase-like 1 |
| chr7_-_107634287 | 0.21 |
ENSRNOT00000093672
ENSRNOT00000087116 |
Sla
|
src-like adaptor |
| chrX_-_158925630 | 0.20 |
ENSRNOT00000073396
|
Ct45a9
|
cancer/testis antigen family 45 member A9 |
| chr10_+_86399827 | 0.20 |
ENSRNOT00000009299
|
Grb7
|
growth factor receptor bound protein 7 |
| chr17_-_43776460 | 0.20 |
ENSRNOT00000089055
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chr1_+_222310920 | 0.20 |
ENSRNOT00000091465
|
Macrod1
|
MACRO domain containing 1 |
| chr12_-_38995570 | 0.20 |
ENSRNOT00000001806
|
Orai1
|
ORAI calcium release-activated calcium modulator 1 |
| chr1_-_89560469 | 0.20 |
ENSRNOT00000079091
|
Scn1b
|
sodium voltage-gated channel beta subunit 1 |
| chr7_-_130350570 | 0.20 |
ENSRNOT00000055805
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr5_+_139963002 | 0.19 |
ENSRNOT00000048506
|
Col9a2
|
collagen type IX alpha 2 chain |
| chr1_+_86938138 | 0.19 |
ENSRNOT00000075601
|
Ccer2
|
coiled-coil glutamate-rich protein 2 |
| chr12_+_24761210 | 0.19 |
ENSRNOT00000002003
|
Cldn4
|
claudin 4 |
| chr1_-_265298797 | 0.18 |
ENSRNOT00000023137
|
Poll
|
DNA polymerase lambda |
| chr10_+_13839956 | 0.18 |
ENSRNOT00000012794
|
Bricd5
|
BRICHOS domain containing 5 |
| chr11_+_72705129 | 0.18 |
ENSRNOT00000073330
|
Apod
|
apolipoprotein D |
| chr8_-_61917125 | 0.18 |
ENSRNOT00000085049
|
RGD1305464
|
similar to human chromosome 15 open reading frame 39 |
| chr19_-_37427989 | 0.18 |
ENSRNOT00000022863
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr2_+_187740531 | 0.17 |
ENSRNOT00000092653
|
Paqr6
|
progestin and adipoQ receptor family member 6 |
| chr1_-_43831932 | 0.17 |
ENSRNOT00000082931
|
Cnksr3
|
Cnksr family member 3 |
| chr15_-_28406046 | 0.17 |
ENSRNOT00000015418
|
Zfp219
|
zinc finger protein 219 |
| chrX_+_134539398 | 0.17 |
ENSRNOT00000004958
|
Cxxc5
|
CXXC finger protein 5 |
| chr20_+_3677474 | 0.16 |
ENSRNOT00000047663
|
Mt1m
|
metallothionein 1M |
| chr15_-_37983882 | 0.16 |
ENSRNOT00000087978
|
Lats2
|
large tumor suppressor kinase 2 |
| chr9_+_80118029 | 0.16 |
ENSRNOT00000023068
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr5_-_134526089 | 0.16 |
ENSRNOT00000013321
|
Cyp4b1
|
cytochrome P450, family 4, subfamily b, polypeptide 1 |
| chr10_-_57243435 | 0.16 |
ENSRNOT00000005050
|
Chrne
|
cholinergic receptor nicotinic epsilon subunit |
| chr4_-_180505916 | 0.16 |
ENSRNOT00000086465
|
AABR07062512.1
|
|
| chr20_+_4966817 | 0.16 |
ENSRNOT00000081527
ENSRNOT00000081265 |
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr17_-_13393243 | 0.16 |
ENSRNOT00000018252
|
Gadd45g
|
growth arrest and DNA-damage-inducible, gamma |
| chr20_+_2004052 | 0.16 |
ENSRNOT00000001008
|
Mog
|
myelin oligodendrocyte glycoprotein |
| chr1_-_8751198 | 0.16 |
ENSRNOT00000030511
|
Adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr16_+_20555395 | 0.16 |
ENSRNOT00000026652
|
Gdf15
|
growth differentiation factor 15 |
| chr5_-_142845116 | 0.15 |
ENSRNOT00000065105
|
RGD1559909
|
RGD1559909 |
| chr1_+_226687258 | 0.15 |
ENSRNOT00000079679
|
Vwce
|
von Willebrand factor C and EGF domains |
| chr13_+_71107465 | 0.15 |
ENSRNOT00000003239
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr1_-_7801438 | 0.15 |
ENSRNOT00000022273
|
AABR07000276.1
|
|
| chr18_+_24708115 | 0.15 |
ENSRNOT00000061054
|
Lims2
|
LIM zinc finger domain containing 2 |
| chr10_-_45534570 | 0.14 |
ENSRNOT00000058362
|
Gjc2
|
gap junction protein, gamma 2 |
| chr4_-_129430251 | 0.14 |
ENSRNOT00000067881
|
Fam19a4
|
family with sequence similarity 19 member A4, C-C motif chemokine like |
| chr3_-_168018410 | 0.14 |
ENSRNOT00000087579
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr14_+_16491573 | 0.14 |
ENSRNOT00000002995
|
Sowahb
|
sosondowah ankyrin repeat domain family member B |
| chr1_+_203524426 | 0.14 |
ENSRNOT00000028020
|
Bub3
|
BUB3 mitotic checkpoint protein |
| chr1_-_89559960 | 0.14 |
ENSRNOT00000092133
|
Scn1b
|
sodium voltage-gated channel beta subunit 1 |
| chr3_+_119014620 | 0.14 |
ENSRNOT00000067700
|
Slc27a2
|
solute carrier family 27 member 2 |
| chr1_-_215846911 | 0.14 |
ENSRNOT00000089171
|
Igf2
|
insulin-like growth factor 2 |
| chr1_+_222311253 | 0.14 |
ENSRNOT00000028749
|
Macrod1
|
MACRO domain containing 1 |
| chr4_-_50860756 | 0.14 |
ENSRNOT00000068404
|
Cadps2
|
calcium dependent secretion activator 2 |
| chr1_+_141550633 | 0.14 |
ENSRNOT00000020047
|
Mesp2
|
mesoderm posterior bHLH transcription factor 2 |
| chr3_+_175144495 | 0.13 |
ENSRNOT00000082601
ENSRNOT00000088026 |
Cdh4
|
cadherin 4 |
| chr19_-_43911057 | 0.13 |
ENSRNOT00000026017
|
Ctrb1
|
chymotrypsinogen B1 |
| chr1_+_98440186 | 0.13 |
ENSRNOT00000024150
|
Iglon5
|
IgLON family member 5 |
| chr15_+_45834302 | 0.13 |
ENSRNOT00000051307
|
Serpine3
|
serpin family E member 3 |
| chr1_+_40529045 | 0.13 |
ENSRNOT00000026564
|
Mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr7_-_107616038 | 0.13 |
ENSRNOT00000088752
|
Sla
|
src-like adaptor |
| chr10_-_38838272 | 0.12 |
ENSRNOT00000089495
|
Sowaha
|
sosondowah ankyrin repeat domain family member A |
| chr10_-_39373437 | 0.12 |
ENSRNOT00000058907
|
Slc22a5
|
solute carrier family 22 member 5 |
| chr20_+_13732198 | 0.12 |
ENSRNOT00000008608
|
LOC103694877
|
macrophage migration inhibitory factor |
| chr16_-_19399851 | 0.12 |
ENSRNOT00000089056
ENSRNOT00000021073 |
Tpm4
|
tropomyosin 4 |
| chr4_-_89151184 | 0.12 |
ENSRNOT00000010263
|
Nap1l5
|
nucleosome assembly protein 1-like 5 |
| chr10_-_88325222 | 0.12 |
ENSRNOT00000021322
ENSRNOT00000076585 |
P3h4
|
prolyl 3-hydroxylase family member 4 (non-enzymatic) |
| chr14_+_66598259 | 0.12 |
ENSRNOT00000049743
|
Kcnip4
|
potassium voltage-gated channel interacting protein 4 |
| chr13_-_52752997 | 0.11 |
ENSRNOT00000013531
|
Pkp1
|
plakophilin 1 |
| chr1_-_242765807 | 0.11 |
ENSRNOT00000020763
|
Pgm5
|
phosphoglucomutase 5 |
| chr17_+_76079720 | 0.11 |
ENSRNOT00000073933
|
Proser2
|
proline and serine rich 2 |
| chr6_-_110904288 | 0.11 |
ENSRNOT00000014645
|
Irf2bpl
|
interferon regulatory factor 2 binding protein-like |
| chr15_-_29360198 | 0.11 |
ENSRNOT00000072745
|
LOC688340
|
T-cell receptor alpha chain V region PHDS58-like |
| chr20_+_5446104 | 0.11 |
ENSRNOT00000000550
|
B3galt4
|
Beta-1,3-galactosyltransferase 4 |
| chr3_+_119015412 | 0.11 |
ENSRNOT00000013605
|
Slc27a2
|
solute carrier family 27 member 2 |
| chr10_+_59799123 | 0.11 |
ENSRNOT00000026493
|
Trpv1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr8_+_75516904 | 0.11 |
ENSRNOT00000013142
|
Rora
|
RAR-related orphan receptor A |
| chr5_-_137372524 | 0.11 |
ENSRNOT00000009061
|
Tmem125
|
transmembrane protein 125 |
| chr15_-_29369504 | 0.11 |
ENSRNOT00000060297
|
AABR07017624.1
|
|
| chr15_+_31243097 | 0.11 |
ENSRNOT00000042721
|
LOC100360891
|
RGD1359684 protein-like |
| chr4_-_157358262 | 0.11 |
ENSRNOT00000021481
|
Gnb3
|
G protein subunit beta 3 |
| chr5_+_151776004 | 0.11 |
ENSRNOT00000009683
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chrX_-_23187341 | 0.11 |
ENSRNOT00000000180
|
Alas2
|
5'-aminolevulinate synthase 2 |
| chr10_+_47490153 | 0.10 |
ENSRNOT00000003182
|
Aldh3a1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr5_-_82168347 | 0.10 |
ENSRNOT00000084959
ENSRNOT00000084147 |
Astn2
|
astrotactin 2 |
| chr2_+_22000106 | 0.10 |
ENSRNOT00000061812
|
Ankrd34b
|
ankyrin repeat domain 34B |
| chr4_-_157358007 | 0.10 |
ENSRNOT00000090898
|
Gnb3
|
G protein subunit beta 3 |
| chr4_-_179700130 | 0.10 |
ENSRNOT00000021306
|
Lmntd1
|
lamin tail domain containing 1 |
| chr10_-_71837851 | 0.10 |
ENSRNOT00000000258
|
Aatf
|
apoptosis antagonizing transcription factor |
| chr1_+_282134981 | 0.10 |
ENSRNOT00000036203
|
Nanos1
|
nanos C2HC-type zinc finger 1 |
| chr4_-_100099517 | 0.10 |
ENSRNOT00000014277
|
Atoh8
|
atonal bHLH transcription factor 8 |
| chr8_-_122841477 | 0.10 |
ENSRNOT00000014861
|
Cmtm7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr11_+_31428358 | 0.10 |
ENSRNOT00000002827
|
Olig1
|
oligodendrocyte transcription factor 1 |
| chr12_+_2180150 | 0.10 |
ENSRNOT00000001322
|
Stxbp2
|
syntaxin binding protein 2 |
| chr10_+_76280933 | 0.10 |
ENSRNOT00000051823
|
RGD1304870
|
similar to Tceb2 protein |
| chr3_+_2648885 | 0.09 |
ENSRNOT00000020339
|
Abca2
|
ATP binding cassette subfamily A member 2 |
| chr10_-_105412627 | 0.09 |
ENSRNOT00000034817
|
Qrich2
|
glutamine rich 2 |
| chr4_+_28972434 | 0.09 |
ENSRNOT00000014219
|
Gngt1
|
G protein subunit gamma transducin 1 |
| chr3_-_176465162 | 0.09 |
ENSRNOT00000048807
|
Nkain4
|
Sodium/potassium transporting ATPase interacting 4 |
| chr1_-_227359809 | 0.09 |
ENSRNOT00000088080
|
1700025F22Rik
|
RIKEN cDNA 1700025F22 gene |
| chr2_-_202816562 | 0.09 |
ENSRNOT00000020401
|
Fam46c
|
family with sequence similarity 46, member C |
| chr13_+_47602692 | 0.09 |
ENSRNOT00000038822
|
Fcmr
|
Fc fragment of IgM receptor |
| chr20_+_4967194 | 0.09 |
ENSRNOT00000070846
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr4_-_132296413 | 0.08 |
ENSRNOT00000052326
|
RGD1560513
|
similar to macrophage migration inhibitory factor |
| chr18_+_54206651 | 0.08 |
ENSRNOT00000073295
|
Chsy3
|
chondroitin sulfate synthase 3 |
| chr10_-_14072230 | 0.08 |
ENSRNOT00000018405
|
Tbl3
|
transducin (beta)-like 3 |
| chr8_-_47494982 | 0.08 |
ENSRNOT00000012170
|
Pou2f3
|
POU class 2 homeobox 3 |
| chr8_+_5993941 | 0.08 |
ENSRNOT00000014065
|
Tmem123
|
transmembrane protein 123 |
| chr9_-_10757720 | 0.08 |
ENSRNOT00000083848
|
Uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr7_-_3315856 | 0.08 |
ENSRNOT00000010035
|
Gdf11
|
growth differentiation factor 11 |
| chr2_-_192381716 | 0.08 |
ENSRNOT00000064950
|
AABR07012291.1
|
|
| chr6_+_51662224 | 0.08 |
ENSRNOT00000060006
|
Ccdc71l
|
coiled-coil domain containing 71-like |
| chr10_-_70220558 | 0.08 |
ENSRNOT00000041389
ENSRNOT00000076398 ENSRNOT00000076596 |
Rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chrX_-_111102464 | 0.08 |
ENSRNOT00000084176
|
Ripply1
|
ripply transcriptional repressor 1 |
| chr10_+_103972888 | 0.08 |
ENSRNOT00000067838
|
Kctd2
|
potassium channel tetramerization domain containing 2 |
| chr8_+_116754178 | 0.08 |
ENSRNOT00000068295
ENSRNOT00000084429 |
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr10_+_104359342 | 0.08 |
ENSRNOT00000006327
|
Tsen54
|
tRNA splicing endonuclease subunit 54 |
| chr12_+_47193964 | 0.08 |
ENSRNOT00000001552
ENSRNOT00000039281 |
Cabp1
|
calcium binding protein 1 |
| chr16_+_20110148 | 0.08 |
ENSRNOT00000080146
ENSRNOT00000025312 |
Jak3
|
Janus kinase 3 |
| chr4_+_144382945 | 0.08 |
ENSRNOT00000007601
|
Cav3
|
caveolin 3 |
| chr12_-_21832813 | 0.08 |
ENSRNOT00000075280
|
Cldn3
|
claudin 3 |
| chr1_-_89369960 | 0.08 |
ENSRNOT00000028545
|
Hamp
|
hepcidin antimicrobial peptide |
| chr9_-_10757954 | 0.08 |
ENSRNOT00000075265
|
Uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr8_-_28208466 | 0.08 |
ENSRNOT00000012247
|
Jam3
|
junctional adhesion molecule 3 |
| chr20_+_14050465 | 0.08 |
ENSRNOT00000089319
ENSRNOT00000074783 |
Lrrc75b
|
leucine rich repeat containing 75B |
| chr4_+_44573264 | 0.08 |
ENSRNOT00000080271
|
Cav2
|
caveolin 2 |
| chr4_+_169161585 | 0.07 |
ENSRNOT00000079785
|
Emp1
|
epithelial membrane protein 1 |
| chr14_-_77810147 | 0.07 |
ENSRNOT00000035427
|
Cytl1
|
cytokine like 1 |
| chr6_+_3657325 | 0.07 |
ENSRNOT00000010927
|
Tmem178a
|
transmembrane protein 178A |
| chr1_-_213987053 | 0.07 |
ENSRNOT00000072774
|
LOC100911519
|
p53-induced protein with a death domain-like |
| chrX_+_6791136 | 0.07 |
ENSRNOT00000003984
|
LOC100909913
|
norrin-like |
| chr2_+_95008311 | 0.07 |
ENSRNOT00000077270
|
Tpd52
|
tumor protein D52 |
| chrX_-_123601100 | 0.07 |
ENSRNOT00000092546
ENSRNOT00000092301 |
Sept6
|
septin 6 |
| chr1_+_83965798 | 0.07 |
ENSRNOT00000068322
|
Cyp2t1
|
cytochrome P450, family 2, subfamily t, polypeptide 1 |
| chr20_-_4380084 | 0.07 |
ENSRNOT00000000499
|
Egfl8
|
EGF-like-domain, multiple 8 |
| chr19_-_11057254 | 0.07 |
ENSRNOT00000025559
|
Herpud1
|
homocysteine inducible ER protein with ubiquitin like domain 1 |
| chr14_-_86395120 | 0.07 |
ENSRNOT00000006802
ENSRNOT00000078572 |
Tmed4
|
transmembrane p24 trafficking protein 4 |
| chr4_+_169147243 | 0.07 |
ENSRNOT00000011580
|
Emp1
|
epithelial membrane protein 1 |
| chr9_-_95290931 | 0.07 |
ENSRNOT00000025300
|
Dnajb3
|
DnaJ heat shock protein family (Hsp40) member B3 |
| chr4_+_157126935 | 0.07 |
ENSRNOT00000056051
|
C1r
|
complement C1r |
| chrX_+_123404518 | 0.07 |
ENSRNOT00000015085
|
Slc25a5
|
solute carrier family 25 member 5 |
| chr13_+_48367307 | 0.06 |
ENSRNOT00000074512
|
LOC100909648
|
vasopressin V1b receptor-like |
| chrX_-_63807810 | 0.06 |
ENSRNOT00000084632
|
Maged1
|
MAGE family member D1 |
| chr2_+_45104305 | 0.06 |
ENSRNOT00000014559
|
Esm1
|
endothelial cell-specific molecule 1 |
| chr8_+_116324040 | 0.06 |
ENSRNOT00000081353
|
Hyal2
|
hyaluronoglucosaminidase 2 |
| chr17_-_72046381 | 0.06 |
ENSRNOT00000073919
|
AABR07028488.1
|
|
| chr18_+_57011575 | 0.06 |
ENSRNOT00000026679
|
Il17b
|
interleukin 17B |
| chr10_+_14216155 | 0.06 |
ENSRNOT00000020192
|
Hagh
|
hydroxyacyl glutathione hydrolase |
| chr19_-_26181449 | 0.06 |
ENSRNOT00000036953
|
LOC685513
|
similar to Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-12 subunit precursor |
| chr12_+_24473981 | 0.06 |
ENSRNOT00000001973
|
Fzd9
|
frizzled class receptor 9 |
| chr4_+_14001761 | 0.06 |
ENSRNOT00000076519
|
Cd36
|
CD36 molecule |
| chr7_+_59349745 | 0.06 |
ENSRNOT00000085334
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr1_-_226935689 | 0.06 |
ENSRNOT00000038807
|
Tmem109
|
transmembrane protein 109 |
| chr15_+_50891127 | 0.06 |
ENSRNOT00000020728
|
Stc1
|
stanniocalcin 1 |
| chr10_+_5352933 | 0.06 |
ENSRNOT00000003465
|
Tekt5
|
tektin 5 |
| chr1_+_218076116 | 0.06 |
ENSRNOT00000028374
|
Oraov1
|
oral cancer overexpressed 1 |
| chr6_-_80334522 | 0.06 |
ENSRNOT00000059316
|
Fbxo33
|
F-box protein 33 |
| chr14_+_13192347 | 0.06 |
ENSRNOT00000000092
|
Antxr2
|
anthrax toxin receptor 2 |
| chr10_-_15204143 | 0.06 |
ENSRNOT00000026971
|
Rhbdl1
|
rhomboid like 1 |
| chr12_-_42492526 | 0.06 |
ENSRNOT00000084018
|
Tbx3
|
T-box 3 |
| chr1_+_100832324 | 0.06 |
ENSRNOT00000056364
|
Il4i1
|
interleukin 4 induced 1 |
| chr12_+_9728486 | 0.06 |
ENSRNOT00000001263
|
Lnx2
|
ligand of numb-protein X 2 |
| chr5_-_164959851 | 0.06 |
ENSRNOT00000012301
|
Fbxo6
|
F-box protein 6 |
| chr2_+_95045034 | 0.06 |
ENSRNOT00000081305
|
Mrps28
|
mitochondrial ribosomal protein S28 |
| chr19_+_37476095 | 0.06 |
ENSRNOT00000092794
ENSRNOT00000023130 |
Hsd11b2
|
hydroxysteroid 11-beta dehydrogenase 2 |
| chr18_+_52215682 | 0.06 |
ENSRNOT00000037901
|
Megf10
|
multiple EGF-like domains 10 |
| chr16_+_53998560 | 0.05 |
ENSRNOT00000077188
ENSRNOT00000013463 |
Asah1
|
N-acylsphingosine amidohydrolase 1 |
| chr18_+_32594958 | 0.05 |
ENSRNOT00000018511
|
Spry4
|
sprouty RTK signaling antagonist 4 |
| chr2_-_30246010 | 0.05 |
ENSRNOT00000023900
|
Mccc2
|
methylcrotonoyl-CoA carboxylase 2 |
| chr12_-_42492328 | 0.05 |
ENSRNOT00000011552
|
Tbx3
|
T-box 3 |
| chr1_+_18491384 | 0.05 |
ENSRNOT00000079138
ENSRNOT00000014917 |
Lama2
|
laminin subunit alpha 2 |
| chr8_+_94243372 | 0.05 |
ENSRNOT00000012864
|
Rwdd2a
|
RWD domain containing 2A |
| chr9_+_69790831 | 0.05 |
ENSRNOT00000045156
|
LOC100360781
|
ribosomal protein L37-like |
| chr3_-_147865393 | 0.05 |
ENSRNOT00000009852
|
Sox12
|
SRY box 12 |
| chr4_+_129574264 | 0.05 |
ENSRNOT00000010185
|
Arl6ip5
|
ADP-ribosylation factor like GTPase 6 interacting protein 5 |
| chr3_+_113415774 | 0.05 |
ENSRNOT00000056151
|
Serf2
|
small EDRK-rich factor 2 |
| chr5_-_59025631 | 0.05 |
ENSRNOT00000049000
ENSRNOT00000022801 |
Tpm2
|
tropomyosin 2, beta |
| chr7_-_2941122 | 0.05 |
ENSRNOT00000082107
|
Esyt1
|
extended synaptotagmin 1 |
| chr10_-_45279299 | 0.05 |
ENSRNOT00000073082
|
Rnf187
|
ring finger protein 187 |
| chr7_-_107768072 | 0.05 |
ENSRNOT00000093189
|
Ndrg1
|
N-myc downstream regulated 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ascl2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.1 | 0.4 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.1 | 0.3 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.1 | 0.4 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.3 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.1 | 0.3 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.2 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.1 | 0.2 | GO:2000097 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.2 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.1 | GO:0015942 | formate metabolic process(GO:0015942) |
| 0.0 | 0.1 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.1 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.0 | 0.0 | GO:0072098 | intermediate mesoderm development(GO:0048389) pattern specification involved in mesonephros development(GO:0061227) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.0 | 0.1 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.0 | 0.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:0015879 | carnitine transport(GO:0015879) antibiotic transport(GO:0042891) |
| 0.0 | 0.6 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.3 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0045210 | negative regulation of dendritic cell cytokine production(GO:0002731) FasL biosynthetic process(GO:0045210) |
| 0.0 | 0.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.1 | GO:1901423 | response to benzene(GO:1901423) |
| 0.0 | 1.0 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.2 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.1 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.1 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.1 | GO:0048105 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.2 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.1 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.3 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.0 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.0 | 0.1 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.1 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.0 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.0 | GO:0046099 | guanine salvage(GO:0006178) GMP catabolic process(GO:0046038) guanine biosynthetic process(GO:0046099) |
| 0.0 | 0.6 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.3 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 0.6 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.1 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.0 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.0 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.2 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.0 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.0 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.0 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.0 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0032173 | septin collar(GO:0032173) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.0 | GO:1990015 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0070820 | cytolytic granule(GO:0044194) tertiary granule(GO:0070820) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.6 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.1 | 0.8 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.3 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.1 | 0.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 1.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) antibiotic transporter activity(GO:0042895) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.2 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.0 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.0 | GO:0016822 | oxaloacetate decarboxylase activity(GO:0008948) hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |