Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
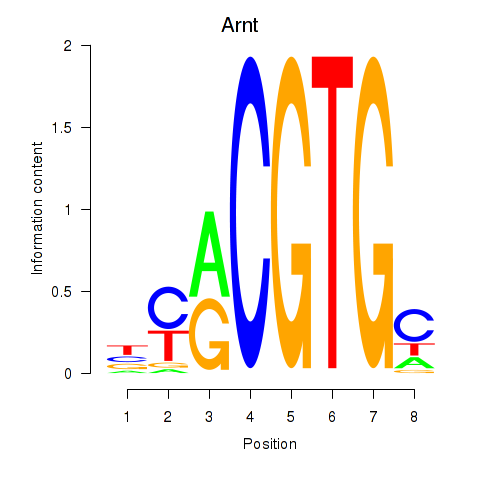
Results for Arnt
Z-value: 0.21
Transcription factors associated with Arnt
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Arnt
|
ENSRNOG00000031174 | aryl hydrocarbon receptor nuclear translocator |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Arnt | rn6_v1_chr2_+_196594303_196594303 | -0.72 | 1.7e-01 | Click! |
Activity profile of Arnt motif
Sorted Z-values of Arnt motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_197991198 | 0.13 |
ENSRNOT00000056322
|
Ciart
|
circadian associated repressor of transcription |
| chr2_-_197991574 | 0.13 |
ENSRNOT00000085632
|
Ciart
|
circadian associated repressor of transcription |
| chr16_+_49462889 | 0.11 |
ENSRNOT00000039909
|
Ankrd37
|
ankyrin repeat domain 37 |
| chr9_-_113331319 | 0.04 |
ENSRNOT00000020681
|
Vapa
|
VAMP associated protein A |
| chr19_+_37226186 | 0.04 |
ENSRNOT00000075933
ENSRNOT00000065013 |
Hsf4
|
heat shock transcription factor 4 |
| chr6_-_26486695 | 0.04 |
ENSRNOT00000073236
|
Krtcap3
|
keratinocyte associated protein 3 |
| chr10_+_16542882 | 0.03 |
ENSRNOT00000028146
|
Stc2
|
stanniocalcin 2 |
| chr5_-_50362344 | 0.03 |
ENSRNOT00000035808
|
Zfp292
|
zinc finger protein 292 |
| chr3_+_113415774 | 0.03 |
ENSRNOT00000056151
|
Serf2
|
small EDRK-rich factor 2 |
| chr4_-_82160240 | 0.02 |
ENSRNOT00000038775
ENSRNOT00000058985 |
Hoxa4
|
homeo box A4 |
| chr2_+_189400696 | 0.02 |
ENSRNOT00000046919
ENSRNOT00000089801 |
LOC361985
|
similar to NICE-3 |
| chr3_-_173953684 | 0.02 |
ENSRNOT00000090468
|
Ppp1r3d
|
protein phosphatase 1, regulatory subunit 3D |
| chr17_+_9837402 | 0.02 |
ENSRNOT00000076436
|
Rab24
|
RAB24, member RAS oncogene family |
| chr13_-_90814119 | 0.02 |
ENSRNOT00000011208
|
Tagln2
|
transgelin 2 |
| chr7_-_18634079 | 0.02 |
ENSRNOT00000010031
|
Angptl4
|
angiopoietin-like 4 |
| chr5_+_141572536 | 0.02 |
ENSRNOT00000023514
|
Rragc
|
Ras-related GTP binding C |
| chr5_+_169506138 | 0.02 |
ENSRNOT00000014904
|
Rpl22
|
ribosomal protein L22 |
| chr4_+_95498003 | 0.02 |
ENSRNOT00000008358
|
Atoh1
|
atonal bHLH transcription factor 1 |
| chr4_-_157679962 | 0.02 |
ENSRNOT00000050443
|
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr1_+_199412834 | 0.02 |
ENSRNOT00000031891
|
Fus
|
fused in sarcoma RNA binding protein |
| chr1_+_154377247 | 0.02 |
ENSRNOT00000092945
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr1_+_261229347 | 0.02 |
ENSRNOT00000018485
|
Ubtd1
|
ubiquitin domain containing 1 |
| chr10_+_69737328 | 0.02 |
ENSRNOT00000055999
ENSRNOT00000076773 |
Tmem132e
|
transmembrane protein 132E |
| chr4_-_117234928 | 0.02 |
ENSRNOT00000073135
|
Pradc1
|
protease-associated domain containing 1 |
| chr11_-_60613718 | 0.02 |
ENSRNOT00000002906
|
Atg3
|
autophagy related 3 |
| chr13_-_51183269 | 0.01 |
ENSRNOT00000039540
|
Ppfia4
|
PTPRF interacting protein alpha 4 |
| chr6_+_99883959 | 0.01 |
ENSRNOT00000010588
|
Fntb
|
farnesyltransferase, CAAX box, beta |
| chr1_+_77994203 | 0.01 |
ENSRNOT00000002044
|
Napa
|
NSF attachment protein alpha |
| chr1_+_220114228 | 0.01 |
ENSRNOT00000026718
|
Ctsf
|
cathepsin F |
| chr8_-_36760742 | 0.01 |
ENSRNOT00000017307
|
Ddx25
|
DEAD-box helicase 25 |
| chr3_+_79678201 | 0.01 |
ENSRNOT00000087604
ENSRNOT00000079709 |
Mtch2
|
mitochondrial carrier 2 |
| chr1_-_93949187 | 0.01 |
ENSRNOT00000018956
|
Zfp536
|
zinc finger protein 536 |
| chr4_-_180234804 | 0.01 |
ENSRNOT00000070957
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr8_-_67869019 | 0.01 |
ENSRNOT00000066009
|
Pias1
|
protein inhibitor of activated STAT, 1 |
| chr7_+_123043503 | 0.01 |
ENSRNOT00000026258
ENSRNOT00000086355 |
Tef
|
TEF, PAR bZIP transcription factor |
| chr4_+_117235023 | 0.01 |
ENSRNOT00000021030
|
Cct7
|
chaperonin containing TCP1 subunit 7 |
| chr10_+_83476107 | 0.01 |
ENSRNOT00000075733
|
Phb
|
prohibitin |
| chr9_+_10428853 | 0.01 |
ENSRNOT00000074253
|
Lonp1
|
lon peptidase 1, mitochondrial |
| chr3_-_159802952 | 0.01 |
ENSRNOT00000011610
|
Oser1
|
oxidative stress responsive serine-rich 1 |
| chr1_+_102849889 | 0.01 |
ENSRNOT00000066791
|
Gtf2h1
|
general transcription factor IIH subunit 1 |
| chr5_-_139227196 | 0.01 |
ENSRNOT00000050941
|
Foxo6
|
forkhead box O6 |
| chr2_+_115337439 | 0.01 |
ENSRNOT00000015779
|
Eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr17_-_9837293 | 0.01 |
ENSRNOT00000091320
ENSRNOT00000022134 |
Prelid1
|
PRELI domain containing 1 |
| chr20_+_5455974 | 0.01 |
ENSRNOT00000000553
ENSRNOT00000092676 |
Pfdn6
|
prefoldin subunit 6 |
| chr9_-_93377643 | 0.01 |
ENSRNOT00000024712
|
Ncl
|
nucleolin |
| chr13_+_70174936 | 0.01 |
ENSRNOT00000064068
ENSRNOT00000079861 ENSRNOT00000092562 |
Arpc5
|
actin related protein 2/3 complex, subunit 5 |
| chr10_-_104097812 | 0.01 |
ENSRNOT00000004867
|
Sumo2
|
small ubiquitin-like modifier 2 |
| chr18_+_1866080 | 0.01 |
ENSRNOT00000018646
|
Snrpd1
|
small nuclear ribonucleoprotein D1 |
| chr4_-_82186190 | 0.01 |
ENSRNOT00000071729
|
LOC100911622
|
homeobox protein Hox-A7-like |
| chr15_-_34338956 | 0.01 |
ENSRNOT00000026914
|
Mdp1
|
magnesium-dependent phosphatase 1 |
| chr15_-_27855999 | 0.01 |
ENSRNOT00000013225
|
Tmem55b
|
transmembrane protein 55B |
| chr8_+_63379087 | 0.00 |
ENSRNOT00000012091
ENSRNOT00000012295 |
Nptn
|
neuroplastin |
| chr4_-_98593664 | 0.00 |
ENSRNOT00000007927
|
Rpia
|
ribose 5-phosphate isomerase A |
| chr20_-_3401273 | 0.00 |
ENSRNOT00000089257
ENSRNOT00000078451 ENSRNOT00000001085 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr1_-_86948845 | 0.00 |
ENSRNOT00000027212
|
Nfkbib
|
NFKB inhibitor beta |
| chr20_-_5455632 | 0.00 |
ENSRNOT00000000552
|
Wdr46
|
WD repeat domain 46 |
| chr6_-_80334522 | 0.00 |
ENSRNOT00000059316
|
Fbxo33
|
F-box protein 33 |
| chr1_-_78573374 | 0.00 |
ENSRNOT00000090519
|
Arhgap35
|
Rho GTPase activating protein 35 |
| chr1_-_129780356 | 0.00 |
ENSRNOT00000077479
|
Arrdc4
|
arrestin domain containing 4 |
| chr5_-_173425611 | 0.00 |
ENSRNOT00000027060
|
B3galt6
|
Beta-1,3-galactosyltransferase 6 |
| chr10_-_35833861 | 0.00 |
ENSRNOT00000049942
|
Canx
|
calnexin |
| chr8_+_50310405 | 0.00 |
ENSRNOT00000073507
|
Sik3
|
SIK family kinase 3 |
| chr1_-_94404211 | 0.00 |
ENSRNOT00000019463
|
Uri1
|
URI1, prefoldin-like chaperone |
| chr4_-_82271893 | 0.00 |
ENSRNOT00000075005
|
Hoxa7
|
homeobox A7 |
| chr1_-_261371508 | 0.00 |
ENSRNOT00000019978
|
Avpi1
|
arginine vasopressin-induced 1 |
| chr9_-_61634510 | 0.00 |
ENSRNOT00000019126
|
Sf3b1
|
splicing factor 3b, subunit 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Arnt
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0033149 | FFAT motif binding(GO:0033149) |