Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
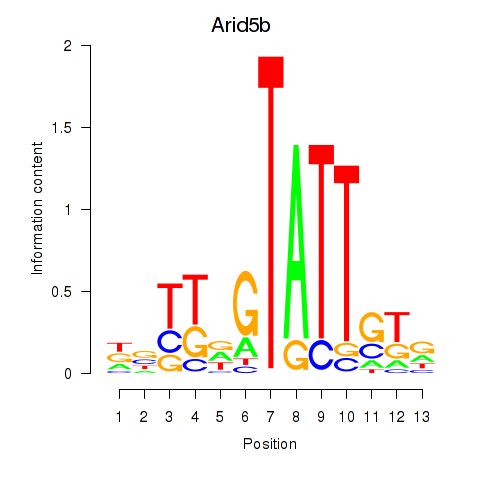
Results for Arid5b
Z-value: 0.45
Transcription factors associated with Arid5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Arid5b
|
ENSRNOG00000000635 | Arid5b |
Activity profile of Arid5b motif
Sorted Z-values of Arid5b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_55696601 | 0.19 |
ENSRNOT00000016127
|
RGD1562914
|
RGD1562914 |
| chr20_+_3558827 | 0.15 |
ENSRNOT00000088130
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr17_+_45199178 | 0.15 |
ENSRNOT00000080047
|
Zscan26
|
zinc finger and SCAN domain containing 26 |
| chr1_-_43638161 | 0.15 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr20_+_4363508 | 0.15 |
ENSRNOT00000077205
|
Ager
|
advanced glycosylation end product-specific receptor |
| chr2_-_208225888 | 0.14 |
ENSRNOT00000054860
|
AABR07012775.1
|
|
| chr4_-_11497531 | 0.13 |
ENSRNOT00000078799
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr11_+_15081774 | 0.13 |
ENSRNOT00000009911
|
LOC108348050
|
ubiquitin carboxyl-terminal hydrolase 25-like |
| chr18_+_14756684 | 0.11 |
ENSRNOT00000076085
ENSRNOT00000076129 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr15_-_39742103 | 0.09 |
ENSRNOT00000074587
ENSRNOT00000036781 ENSRNOT00000071919 |
Setdb2
|
SET domain, bifurcated 2 |
| chr9_-_79898912 | 0.09 |
ENSRNOT00000022076
|
March4
|
membrane associated ring-CH-type finger 4 |
| chr3_+_148448732 | 0.09 |
ENSRNOT00000083812
|
Ttll9
|
tubulin tyrosine ligase like 9 |
| chr19_-_55510460 | 0.09 |
ENSRNOT00000019820
|
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr15_-_9086282 | 0.08 |
ENSRNOT00000008989
|
Thrb
|
thyroid hormone receptor beta |
| chr4_+_140377565 | 0.08 |
ENSRNOT00000082723
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr6_-_42630983 | 0.08 |
ENSRNOT00000071977
|
Atp6v1c2
|
ATPase H+ transporting V1 subunit C2 |
| chr1_+_167051209 | 0.08 |
ENSRNOT00000055249
|
Numa1
|
nuclear mitotic apparatus protein 1 |
| chr5_-_16140896 | 0.08 |
ENSRNOT00000029503
|
Xkr4
|
XK related 4 |
| chr10_-_1744647 | 0.08 |
ENSRNOT00000081886
|
AABR07028997.1
|
|
| chr13_+_90533365 | 0.07 |
ENSRNOT00000082469
|
Dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr4_+_102426224 | 0.07 |
ENSRNOT00000073768
|
LOC103694107
|
NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3 pseudogene |
| chr4_-_55398941 | 0.06 |
ENSRNOT00000075324
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr4_-_165456677 | 0.06 |
ENSRNOT00000082207
|
Klra2
|
killer cell lectin-like receptor, subfamily A, member 2 |
| chr2_-_113345577 | 0.06 |
ENSRNOT00000034096
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr6_-_2961510 | 0.06 |
ENSRNOT00000090688
|
Dhx57
|
DExH-box helicase 57 |
| chr2_+_22950018 | 0.06 |
ENSRNOT00000071804
|
Homer1
|
homer scaffolding protein 1 |
| chr14_+_69800156 | 0.06 |
ENSRNOT00000072746
|
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr10_-_62664466 | 0.06 |
ENSRNOT00000078730
|
Git1
|
GIT ArfGAP 1 |
| chr1_-_167884690 | 0.06 |
ENSRNOT00000091372
|
Olr61
|
olfactory receptor 61 |
| chr2_+_186980992 | 0.05 |
ENSRNOT00000020717
|
Arhgef11
|
Rho guanine nucleotide exchange factor 11 |
| chr4_-_28310178 | 0.05 |
ENSRNOT00000084021
|
RGD1563091
|
similar to OEF2 |
| chr17_-_17947777 | 0.05 |
ENSRNOT00000036876
|
Rnf144b
|
ring finger protein 144B |
| chr1_-_100969560 | 0.05 |
ENSRNOT00000035908
|
Cpt1c
|
carnitine palmitoyltransferase 1c |
| chr2_-_122646445 | 0.05 |
ENSRNOT00000080100
|
Dcun1d1
|
defective in cullin neddylation 1 domain containing 1 |
| chr3_-_105663457 | 0.05 |
ENSRNOT00000067646
|
Zfp770
|
zinc finger protein 770 |
| chr10_-_52290657 | 0.05 |
ENSRNOT00000005293
|
Map2k4
|
mitogen activated protein kinase kinase 4 |
| chr19_-_54648227 | 0.05 |
ENSRNOT00000025424
|
Klhdc4
|
kelch domain containing 4 |
| chr7_+_132378273 | 0.05 |
ENSRNOT00000010990
|
LOC690142
|
hypothetical protein LOC690142 |
| chr10_+_13145099 | 0.05 |
ENSRNOT00000091287
|
Srrm2
|
serine/arginine repetitive matrix 2 |
| chr18_+_30574627 | 0.05 |
ENSRNOT00000060484
|
Pcdhb19
|
protocadherin beta 19 |
| chr8_-_62616828 | 0.05 |
ENSRNOT00000068340
|
Arid3b
|
AT-rich interaction domain 3B |
| chr16_-_69280109 | 0.04 |
ENSRNOT00000058595
|
AABR07026240.1
|
|
| chr15_-_61648267 | 0.04 |
ENSRNOT00000071805
|
Naa16
|
N(alpha)-acetyltransferase 16, NatA auxiliary subunit |
| chr5_+_160009422 | 0.04 |
ENSRNOT00000088644
|
Zbtb17
|
zinc finger and BTB domain containing 17 |
| chr4_+_123760743 | 0.04 |
ENSRNOT00000013498
|
Ccdc174
|
coiled-coil domain containing 174 |
| chr18_-_1828606 | 0.04 |
ENSRNOT00000033312
|
Esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr18_+_17550350 | 0.04 |
ENSRNOT00000078870
|
RGD1562608
|
similar to KIAA1328 protein |
| chr3_-_72081079 | 0.04 |
ENSRNOT00000007914
|
Tmx2
|
thioredoxin-related transmembrane protein 2 |
| chr1_-_225948626 | 0.04 |
ENSRNOT00000077823
|
Incenp
|
inner centromere protein |
| chr7_-_12874215 | 0.04 |
ENSRNOT00000011837
|
Hcn2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2 |
| chr12_-_17311112 | 0.04 |
ENSRNOT00000001732
|
Gper1
|
G protein-coupled estrogen receptor 1 |
| chr6_-_26680284 | 0.04 |
ENSRNOT00000039709
|
Cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr4_-_67301102 | 0.03 |
ENSRNOT00000034549
|
Dennd2a
|
DENN domain containing 2A |
| chr5_-_117612123 | 0.03 |
ENSRNOT00000065112
|
Dock7
|
dedicator of cytokinesis 7 |
| chr1_-_157461588 | 0.03 |
ENSRNOT00000068402
|
Ankrd42
|
ankyrin repeat domain 42 |
| chr8_-_21481735 | 0.03 |
ENSRNOT00000038069
|
Zfp426
|
zinc finger protein 426 |
| chr2_-_258932200 | 0.03 |
ENSRNOT00000045905
|
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr18_-_81428971 | 0.03 |
ENSRNOT00000065201
|
Zfp407
|
zinc finger protein 407 |
| chr3_-_13525983 | 0.03 |
ENSRNOT00000082036
|
Pbx3
|
PBX homeobox 3 |
| chr10_+_71322145 | 0.03 |
ENSRNOT00000079418
|
Synrg
|
synergin, gamma |
| chr14_-_66978499 | 0.03 |
ENSRNOT00000081601
|
Slit2
|
slit guidance ligand 2 |
| chr18_+_60392376 | 0.03 |
ENSRNOT00000023890
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr2_+_226825635 | 0.03 |
ENSRNOT00000042173
|
AABR07013200.1
|
|
| chr1_+_170205591 | 0.02 |
ENSRNOT00000071063
|
LOC686660
|
similar to olfactory receptor 692 |
| chr2_+_185590986 | 0.02 |
ENSRNOT00000088807
ENSRNOT00000088188 |
Lrba
|
LPS responsive beige-like anchor protein |
| chr12_-_24346918 | 0.02 |
ENSRNOT00000001970
|
Nsun5
|
NOP2/Sun RNA methyltransferase family member 5 |
| chr2_+_94266213 | 0.02 |
ENSRNOT00000090023
|
Pag1
|
phosphoprotein membrane anchor with glycosphingolipid microdomains 1 |
| chr2_+_186980793 | 0.02 |
ENSRNOT00000091336
|
Arhgef11
|
Rho guanine nucleotide exchange factor 11 |
| chr17_+_34704616 | 0.02 |
ENSRNOT00000090706
ENSRNOT00000083674 ENSRNOT00000077110 |
Exoc2
|
exocyst complex component 2 |
| chr2_+_83393282 | 0.02 |
ENSRNOT00000044871
|
Ctnnd2
|
catenin delta 2 |
| chr11_+_81757983 | 0.02 |
ENSRNOT00000088503
|
Tbccd1
|
TBCC domain containing 1 |
| chr2_-_26699333 | 0.02 |
ENSRNOT00000024459
|
Sv2c
|
synaptic vesicle glycoprotein 2c |
| chr3_+_72080630 | 0.01 |
ENSRNOT00000008911
|
Med19
|
mediator complex subunit 19 |
| chr8_-_119889661 | 0.01 |
ENSRNOT00000011780
|
Stac
|
SH3 and cysteine rich domain |
| chr1_+_153861948 | 0.01 |
ENSRNOT00000087067
|
Me3
|
malic enzyme 3 |
| chr6_-_88917070 | 0.01 |
ENSRNOT00000000767
|
Mdga2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr4_+_34237762 | 0.01 |
ENSRNOT00000084163
|
Mios
|
meiosis regulator for oocyte development |
| chr8_-_70436028 | 0.01 |
ENSRNOT00000077152
|
Slc24a1
|
solute carrier family 24 member 1 |
| chr10_+_35872619 | 0.01 |
ENSRNOT00000059190
|
Hnrnph1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chr3_-_46726946 | 0.01 |
ENSRNOT00000011030
ENSRNOT00000086576 |
Itgb6
|
integrin subunit beta 6 |
| chr10_-_98469799 | 0.01 |
ENSRNOT00000087502
ENSRNOT00000088646 |
Abca9
|
ATP binding cassette subfamily A member 9 |
| chr5_+_58995249 | 0.01 |
ENSRNOT00000023411
|
Ccdc107
|
coiled-coil domain containing 107 |
| chr17_+_47721977 | 0.01 |
ENSRNOT00000080800
|
LOC100910792
|
amphiphysin-like |
| chr2_-_9023104 | 0.00 |
ENSRNOT00000039652
|
Adgrv1
|
adhesion G protein-coupled receptor V1 |
| chr6_+_123897956 | 0.00 |
ENSRNOT00000005167
|
Tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr20_+_11655967 | 0.00 |
ENSRNOT00000064565
|
LOC690478
|
similar to keratin associated protein 10-7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Arid5b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1905204 | response to selenite ion(GO:0072714) negative regulation of connective tissue replacement(GO:1905204) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.0 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.0 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.0 | 0.1 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.0 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.1 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |