Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
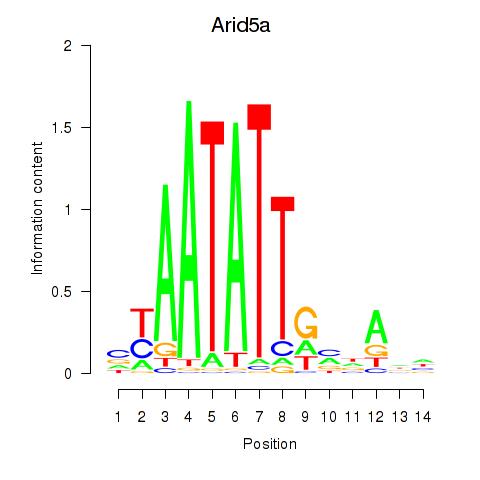
Results for Arid5a
Z-value: 0.11
Transcription factors associated with Arid5a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Arid5a
|
ENSRNOG00000015382 | AT-rich interaction domain 5A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Arid5a | rn6_v1_chr9_+_42871950_42871950 | -0.19 | 7.7e-01 | Click! |
Activity profile of Arid5a motif
Sorted Z-values of Arid5a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_162357356 | 0.04 |
ENSRNOT00000030119
|
Eya2
|
EYA transcriptional coactivator and phosphatase 2 |
| chr14_-_41786084 | 0.03 |
ENSRNOT00000059439
|
Grxcr1
|
glutaredoxin and cysteine rich domain containing 1 |
| chr6_+_123034304 | 0.02 |
ENSRNOT00000078938
|
Cpg1
|
candidate plasticity gene 1 |
| chr13_+_27465930 | 0.02 |
ENSRNOT00000003314
|
Serpinb10
|
serpin family B member 10 |
| chr20_-_5037022 | 0.01 |
ENSRNOT00000091022
ENSRNOT00000060762 |
Msh5
|
mutS homolog 5 |
| chr3_+_146981984 | 0.01 |
ENSRNOT00000090211
|
Nsfl1c
|
NSFL1 cofactor |
| chr13_-_90443157 | 0.01 |
ENSRNOT00000006862
|
Nhlh1
|
nescient helix loop helix 1 |
| chr20_+_17750744 | 0.00 |
ENSRNOT00000000745
|
RGD1559903
|
similar to RIKEN cDNA 1700049L16 |
| chr17_+_43734461 | 0.00 |
ENSRNOT00000072564
|
Hist1h1d
|
histone cluster 1, H1d |
| chr1_-_170431073 | 0.00 |
ENSRNOT00000024710
|
Hpx
|
hemopexin |
| chr5_+_15043955 | 0.00 |
ENSRNOT00000047093
|
Rp1
|
retinitis pigmentosa 1 |
| chr3_-_48831417 | 0.00 |
ENSRNOT00000009920
ENSRNOT00000085246 |
Kcnh7
|
potassium voltage-gated channel subfamily H member 7 |
| chr17_-_42267079 | 0.00 |
ENSRNOT00000059564
|
LOC100911664
|
uncharacterized LOC100911664 |
| chrX_-_23144324 | 0.00 |
ENSRNOT00000000178
ENSRNOT00000081239 |
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |