Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Arid3a
Z-value: 0.17
Transcription factors associated with Arid3a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Arid3a
|
ENSRNOG00000026435 | AT-rich interaction domain 3A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Arid3a | rn6_v1_chr7_-_12598370_12598370 | 0.67 | 2.1e-01 | Click! |
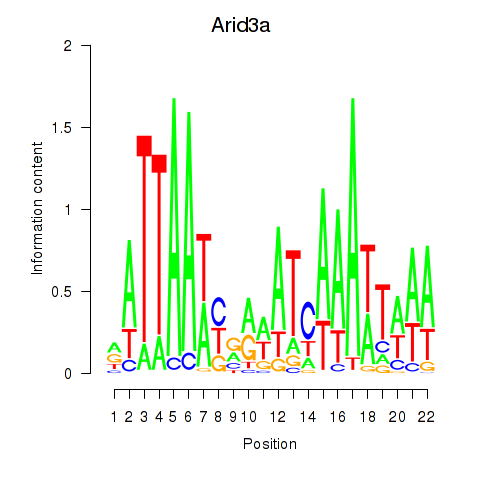
Activity profile of Arid3a motif
Sorted Z-values of Arid3a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_84827062 | 0.09 |
ENSRNOT00000058006
|
A930003A15Rik
|
RIKEN cDNA A930003A15 gene |
| chr9_-_66019065 | 0.09 |
ENSRNOT00000088729
|
Als2
|
ALS2, alsin Rho guanine nucleotide exchange factor |
| chrX_-_40086870 | 0.08 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr3_-_66279155 | 0.06 |
ENSRNOT00000079887
|
Cerkl
|
ceramide kinase-like |
| chr1_-_190370499 | 0.06 |
ENSRNOT00000084389
|
AABR07005618.1
|
|
| chr10_-_59883839 | 0.05 |
ENSRNOT00000093579
|
Aspa
|
aspartoacylase |
| chr10_-_103848035 | 0.04 |
ENSRNOT00000029001
|
Fads6
|
fatty acid desaturase 6 |
| chr6_-_126710854 | 0.04 |
ENSRNOT00000081127
ENSRNOT00000064914 |
Btbd7
|
BTB domain containing 7 |
| chr15_-_110046687 | 0.04 |
ENSRNOT00000057404
ENSRNOT00000006624 ENSRNOT00000089695 |
Nalcn
|
sodium leak channel, non-selective |
| chr13_-_34251942 | 0.04 |
ENSRNOT00000044095
|
Tsn
|
translin |
| chr5_-_128333805 | 0.03 |
ENSRNOT00000037523
|
Zfyve9
|
zinc finger FYVE-type containing 9 |
| chr1_+_87224677 | 0.03 |
ENSRNOT00000028070
|
Ppp1r14a
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr3_+_150686638 | 0.03 |
ENSRNOT00000078235
|
Itch
|
itchy E3 ubiquitin protein ligase |
| chr4_-_98305173 | 0.03 |
ENSRNOT00000010151
|
Il23r
|
interleukin 23 receptor |
| chr18_+_16146447 | 0.03 |
ENSRNOT00000022117
|
Galnt1
|
polypeptide N-acetylgalactosaminyltransferase 1 |
| chr1_+_204959174 | 0.03 |
ENSRNOT00000023257
|
Zranb1
|
zinc finger RANBP2-type containing 1 |
| chr2_-_210454737 | 0.03 |
ENSRNOT00000079993
|
Ahcyl1
|
adenosylhomocysteinase-like 1 |
| chr1_-_189182306 | 0.02 |
ENSRNOT00000021249
|
Gp2
|
glycoprotein 2 |
| chr11_-_71135493 | 0.02 |
ENSRNOT00000050535
|
Fyttd1
|
forty-two-three domain containing 1 |
| chr15_-_4055539 | 0.02 |
ENSRNOT00000090352
|
Sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr6_-_136436818 | 0.02 |
ENSRNOT00000082600
|
Ppp1r13b
|
protein phosphatase 1, regulatory subunit 13B |
| chr13_+_98231326 | 0.02 |
ENSRNOT00000003837
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr7_+_116671948 | 0.02 |
ENSRNOT00000077773
ENSRNOT00000029711 |
Gli4
|
GLI family zinc finger 4 |
| chr14_-_86333424 | 0.02 |
ENSRNOT00000083191
|
Nudcd3
|
NudC domain containing 3 |
| chr8_-_124399494 | 0.02 |
ENSRNOT00000037883
|
Tgfbr2
|
transforming growth factor, beta receptor 2 |
| chr20_-_5123073 | 0.02 |
ENSRNOT00000001126
|
Apom
|
apolipoprotein M |
| chrX_-_63182385 | 0.02 |
ENSRNOT00000076613
|
Zfx
|
zinc finger protein X-linked |
| chr1_+_156552328 | 0.02 |
ENSRNOT00000055401
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr8_+_70708873 | 0.02 |
ENSRNOT00000045106
|
Parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr3_+_48096954 | 0.02 |
ENSRNOT00000068238
ENSRNOT00000064344 ENSRNOT00000044638 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr6_-_51018050 | 0.02 |
ENSRNOT00000082691
|
Gpr22
|
G protein-coupled receptor 22 |
| chr6_-_107325345 | 0.02 |
ENSRNOT00000049481
ENSRNOT00000042594 ENSRNOT00000013026 |
Numb
|
NUMB, endocytic adaptor protein |
| chr8_-_93390305 | 0.02 |
ENSRNOT00000056930
|
Ibtk
|
inhibitor of Bruton tyrosine kinase |
| chr13_+_98311827 | 0.02 |
ENSRNOT00000082844
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr2_-_210116038 | 0.02 |
ENSRNOT00000074950
|
LOC684509
|
similar to NADH-ubiquinone oxidoreductase B9 subunit (Complex I-B9) (CI-B9) |
| chr12_-_38916237 | 0.02 |
ENSRNOT00000074517
|
Tmem120b
|
transmembrane protein 120B |
| chr12_-_1195591 | 0.02 |
ENSRNOT00000001446
|
Stard13
|
StAR-related lipid transfer domain containing 13 |
| chr9_-_65442257 | 0.02 |
ENSRNOT00000037660
|
Fam126b
|
family with sequence similarity 126, member B |
| chr1_-_77844189 | 0.02 |
ENSRNOT00000017555
|
Gltscr2
|
glioma tumor suppressor candidate region gene 2 |
| chr14_+_39964588 | 0.02 |
ENSRNOT00000003240
|
Gabrg1
|
gamma-aminobutyric acid type A receptor gamma 1 subunit |
| chr4_-_155690869 | 0.02 |
ENSRNOT00000012216
|
C3ar1
|
complement C3a receptor 1 |
| chr4_-_81241152 | 0.02 |
ENSRNOT00000015325
ENSRNOT00000015152 |
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr17_+_84881414 | 0.02 |
ENSRNOT00000034157
|
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr1_+_95397991 | 0.02 |
ENSRNOT00000039649
|
Zfp939
|
zinc finger protein 939 |
| chr11_-_60547201 | 0.02 |
ENSRNOT00000093151
|
Btla
|
B and T lymphocyte associated |
| chr14_-_28967980 | 0.02 |
ENSRNOT00000048175
|
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr5_-_12199283 | 0.02 |
ENSRNOT00000007769
|
Pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr1_-_189181901 | 0.02 |
ENSRNOT00000092022
|
Gp2
|
glycoprotein 2 |
| chr1_+_99749936 | 0.02 |
ENSRNOT00000025299
|
Klk7
|
kallikrein-related peptidase 7 |
| chr14_-_44078897 | 0.02 |
ENSRNOT00000031792
|
N4bp2
|
NEDD4 binding protein 2 |
| chr7_-_63045728 | 0.02 |
ENSRNOT00000039532
|
Lemd3
|
LEM domain containing 3 |
| chr19_+_55206455 | 0.02 |
ENSRNOT00000039273
|
Zc3h18
|
zinc finger CCCH-type containing 18 |
| chr14_+_104394200 | 0.01 |
ENSRNOT00000088356
|
AABR07016566.1
|
|
| chr12_-_21746236 | 0.01 |
ENSRNOT00000001869
|
LOC102550456
|
TSC22 domain family protein 4-like |
| chr9_+_77834091 | 0.01 |
ENSRNOT00000033459
|
Vwc2l
|
von Willebrand factor C domain-containing protein 2-like |
| chr4_-_161658519 | 0.01 |
ENSRNOT00000007634
ENSRNOT00000067895 |
Tulp3
|
tubby-like protein 3 |
| chr5_+_36566783 | 0.01 |
ENSRNOT00000077650
|
Fbxl4
|
F-box and leucine-rich repeat protein 4 |
| chr3_-_64766472 | 0.01 |
ENSRNOT00000037684
|
Cwc22
|
CWC22 spliceosome associated protein homolog |
| chr8_+_59420123 | 0.01 |
ENSRNOT00000077922
|
Ireb2
|
iron responsive element binding protein 2 |
| chr10_+_97771264 | 0.01 |
ENSRNOT00000005257
|
Arsg
|
arylsulfatase G |
| chr1_+_64740487 | 0.01 |
ENSRNOT00000081213
|
LOC103691005
|
zinc finger protein 679-like |
| chr20_+_27212724 | 0.01 |
ENSRNOT00000032600
|
Hnrnph3
|
heterogeneous nuclear ribonucleoprotein H3 |
| chr11_+_20474483 | 0.01 |
ENSRNOT00000082417
ENSRNOT00000002895 |
Ncam2
|
neural cell adhesion molecule 2 |
| chr15_-_6587367 | 0.01 |
ENSRNOT00000038449
|
Zfp385d
|
zinc finger protein 385D |
| chr13_+_50873605 | 0.01 |
ENSRNOT00000004382
|
Fmod
|
fibromodulin |
| chr10_+_61640015 | 0.01 |
ENSRNOT00000092714
|
Mettl16
|
methyltransferase like 16 |
| chr1_-_185569190 | 0.01 |
ENSRNOT00000090773
|
RGD1311703
|
similar to sid2057p |
| chr4_+_5841998 | 0.01 |
ENSRNOT00000010025
|
Xrcc2
|
X-ray repair cross complementing 2 |
| chr8_-_114010277 | 0.01 |
ENSRNOT00000045087
|
Atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr17_+_1305016 | 0.01 |
ENSRNOT00000025965
|
Ercc6l2
|
ERCC excision repair 6 like 2 |
| chr1_+_68436917 | 0.01 |
ENSRNOT00000088586
|
LOC108348118
|
leucyl-cystinyl aminopeptidase |
| chr9_+_111597037 | 0.01 |
ENSRNOT00000021758
|
Fer
|
FER tyrosine kinase |
| chr18_+_30840868 | 0.01 |
ENSRNOT00000027026
|
Pcdhga5
|
protocadherin gamma subfamily A, 5 |
| chr3_+_47677720 | 0.01 |
ENSRNOT00000065340
|
Tbr1
|
T-box, brain, 1 |
| chr12_+_51960262 | 0.01 |
ENSRNOT00000084214
|
Ep400
|
E1A binding protein p400 |
| chr1_+_68436593 | 0.01 |
ENSRNOT00000080325
|
LOC108348118
|
leucyl-cystinyl aminopeptidase |
| chr10_-_11877584 | 0.01 |
ENSRNOT00000078577
|
Cluap1
|
clusterin associated protein 1 |
| chr11_-_4332255 | 0.01 |
ENSRNOT00000087133
|
Cadm2
|
cell adhesion molecule 2 |
| chr11_-_72109964 | 0.01 |
ENSRNOT00000058917
|
AABR07034445.1
|
|
| chr8_+_70760922 | 0.01 |
ENSRNOT00000044887
|
Cilp
|
cartilage intermediate layer protein |
| chr6_-_146470456 | 0.01 |
ENSRNOT00000018479
|
RGD1560883
|
similar to KIAA0825 protein |
| chr7_-_12601674 | 0.01 |
ENSRNOT00000093489
|
Arid3a
|
AT-rich interaction domain 3A |
| chr16_+_8737974 | 0.01 |
ENSRNOT00000064255
|
Ercc6
|
ERCC excision repair 6, chromatin remodeling factor |
| chr8_+_103938520 | 0.01 |
ENSRNOT00000067835
|
Gk5
|
glycerol kinase 5 (putative) |
| chr2_+_186703541 | 0.01 |
ENSRNOT00000093342
|
Fcrl1
|
Fc receptor-like 1 |
| chr9_-_5329305 | 0.01 |
ENSRNOT00000078055
|
Slc5a7
|
solute carrier family 5 member 7 |
| chr16_-_14382641 | 0.01 |
ENSRNOT00000018723
|
Ghitm
|
growth hormone inducible transmembrane protein |
| chr12_-_40332612 | 0.01 |
ENSRNOT00000001691
|
Atxn2
|
ataxin 2 |
| chr10_+_76320041 | 0.01 |
ENSRNOT00000037693
|
Coil
|
coilin |
| chr5_-_126323799 | 0.01 |
ENSRNOT00000089934
|
Mroh7
|
maestro heat-like repeat family member 7 |
| chr6_-_108120579 | 0.00 |
ENSRNOT00000041163
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr16_-_80850340 | 0.00 |
ENSRNOT00000000124
|
Champ1
|
chromosome alignment maintaining phosphoprotein 1 |
| chrX_-_105417323 | 0.00 |
ENSRNOT00000015494
|
Gla
|
galactosidase, alpha |
| chr3_+_63510293 | 0.00 |
ENSRNOT00000058093
|
Dfnb59
|
deafness, autosomal recessive 59 |
| chr7_-_27214236 | 0.00 |
ENSRNOT00000034213
|
Tdg
|
thymine-DNA glycosylase |
| chr5_-_25721072 | 0.00 |
ENSRNOT00000021839
|
Tmem67
|
transmembrane protein 67 |
| chr8_-_116993193 | 0.00 |
ENSRNOT00000026327
|
Dag1
|
dystroglycan 1 |
| chr5_+_120340646 | 0.00 |
ENSRNOT00000086259
ENSRNOT00000086539 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr9_+_42945358 | 0.00 |
ENSRNOT00000059806
|
Fer1l5
|
fer-1-like family member 5 |
| chr1_+_61268248 | 0.00 |
ENSRNOT00000082730
|
LOC102552527
|
zinc finger protein 420-like |
| chr12_-_29958050 | 0.00 |
ENSRNOT00000058725
|
Tmem248
|
transmembrane protein 248 |
| chr14_+_100415668 | 0.00 |
ENSRNOT00000008057
|
C1d
|
C1D nuclear receptor co-repressor |
| chr12_+_46869836 | 0.00 |
ENSRNOT00000084421
|
Sirt4
|
sirtuin 4 |
| chr18_-_31749647 | 0.00 |
ENSRNOT00000044287
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr5_+_103251986 | 0.00 |
ENSRNOT00000008757
|
Cntln
|
centlein |
| chr8_-_109621408 | 0.00 |
ENSRNOT00000087398
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr12_-_29919320 | 0.00 |
ENSRNOT00000038092
|
Tyw1
|
tRNA-yW synthesizing protein 1 homolog |
| chr6_+_99444013 | 0.00 |
ENSRNOT00000058642
|
Ppp1r36
|
protein phosphatase 1, regulatory subunit 36 |
| chr8_+_82038967 | 0.00 |
ENSRNOT00000079535
|
Myo5a
|
myosin VA |
| chr7_-_12432130 | 0.00 |
ENSRNOT00000077301
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr2_+_189857587 | 0.00 |
ENSRNOT00000048214
|
Ilf2
|
interleukin enhancer binding factor 2 |
| chr4_+_61850348 | 0.00 |
ENSRNOT00000013423
|
Akr1b7
|
aldo-keto reductase family 1, member B7 |
| chr16_+_70644474 | 0.00 |
ENSRNOT00000045955
|
LOC100359503
|
ribosomal protein S28-like |
| chrX_-_64726210 | 0.00 |
ENSRNOT00000076012
ENSRNOT00000086265 |
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr2_+_52397175 | 0.00 |
ENSRNOT00000082614
|
Ccl28
|
C-C motif chemokine ligand 28 |
| chr17_-_53713408 | 0.00 |
ENSRNOT00000022445
|
LOC100912163
|
AT-rich interactive domain-containing protein 4B-like |
| chr19_-_44095250 | 0.00 |
ENSRNOT00000074824
|
Tmem170a
|
transmembrane protein 170A |
| chr9_-_86103158 | 0.00 |
ENSRNOT00000021528
|
Cul3
|
cullin 3 |
| chr17_-_53915076 | 0.00 |
ENSRNOT00000010836
|
Arid4b
|
AT-rich interaction domain 4B |
| chr8_-_77599781 | 0.00 |
ENSRNOT00000087980
|
Aqp9
|
aquaporin 9 |
| chr3_+_95939260 | 0.00 |
ENSRNOT00000041291
|
AABR07053188.1
|
|
| chr17_+_54181419 | 0.00 |
ENSRNOT00000023861
|
Kif5b
|
kinesin family member 5B |
| chr10_+_65272849 | 0.00 |
ENSRNOT00000014386
|
Eral1
|
Era-like 12S mitochondrial rRNA chaperone 1 |
| chr2_+_128675814 | 0.00 |
ENSRNOT00000058366
|
RGD1359508
|
similar to protein C33A12.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Arid3a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.0 | 0.1 | GO:0051036 | regulation of endosome size(GO:0051036) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.0 | 0.1 | GO:0001729 | ceramide kinase activity(GO:0001729) |