Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
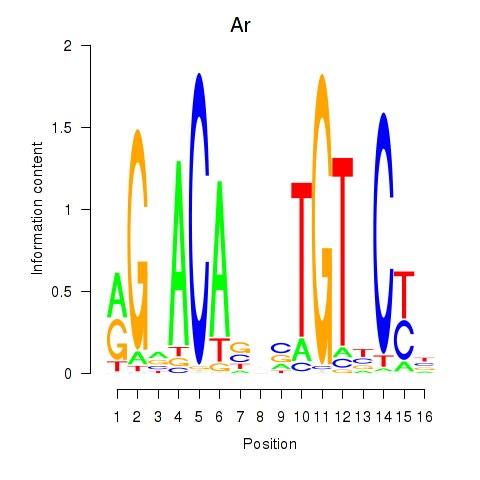
Results for Ar
Z-value: 1.40
Transcription factors associated with Ar
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ar
|
ENSRNOG00000005639 | androgen receptor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ar | rn6_v1_chrX_+_67656253_67656253 | 0.32 | 5.9e-01 | Click! |
Activity profile of Ar motif
Sorted Z-values of Ar motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_27603307 | 0.98 |
ENSRNOT00000074702
|
AABR07043347.1
|
|
| chr15_-_42271443 | 0.96 |
ENSRNOT00000085508
|
AABR07018132.1
|
|
| chr15_+_64939527 | 0.93 |
ENSRNOT00000045865
|
AABR07018556.1
|
|
| chr2_+_89254154 | 0.90 |
ENSRNOT00000073391
|
AABR07009221.1
|
|
| chr3_+_116072294 | 0.89 |
ENSRNOT00000071786
|
AABR07053613.1
|
|
| chr2_-_167607919 | 0.88 |
ENSRNOT00000089083
|
AABR07011733.1
|
|
| chr2_+_52152536 | 0.87 |
ENSRNOT00000072691
|
AABR07008293.2
|
|
| chr19_+_24655626 | 0.87 |
ENSRNOT00000086690
|
AC102976.1
|
|
| chr3_-_76012929 | 0.87 |
ENSRNOT00000085199
|
AC111632.2
|
|
| chr6_+_54438436 | 0.86 |
ENSRNOT00000091972
|
AABR07063893.1
|
|
| chr3_+_95614562 | 0.86 |
ENSRNOT00000079990
|
AABR07053179.1
|
|
| chr5_-_79899054 | 0.85 |
ENSRNOT00000074379
|
AC229945.1
|
|
| chr14_-_43855745 | 0.85 |
ENSRNOT00000083155
|
AC112624.1
|
|
| chrX_+_153677811 | 0.85 |
ENSRNOT00000072808
|
AABR07042361.1
|
|
| chr11_-_17286846 | 0.84 |
ENSRNOT00000080073
|
AABR07033318.1
|
|
| chr7_+_96195209 | 0.83 |
ENSRNOT00000073584
|
AABR07057961.1
|
|
| chr10_-_52483325 | 0.82 |
ENSRNOT00000083485
|
AABR07029809.1
|
|
| chr13_+_29839867 | 0.81 |
ENSRNOT00000090623
|
AABR07020537.1
|
|
| chr19_-_21970103 | 0.81 |
ENSRNOT00000074210
|
AABR07043115.1
|
|
| chr6_-_86161333 | 0.79 |
ENSRNOT00000072471
|
AABR07064590.1
|
|
| chr16_+_33373423 | 0.77 |
ENSRNOT00000091672
|
AABR07025316.1
|
|
| chr16_+_33373615 | 0.75 |
ENSRNOT00000079157
|
AABR07025316.1
|
|
| chr2_+_166475070 | 0.75 |
ENSRNOT00000071111
|
AABR07011700.1
|
|
| chr2_+_70490155 | 0.72 |
ENSRNOT00000078674
|
AABR07008681.1
|
|
| chrX_+_82710329 | 0.68 |
ENSRNOT00000074962
|
AABR07039694.1
|
|
| chr3_-_8432593 | 0.67 |
ENSRNOT00000090574
|
AC114363.1
|
|
| chr8_-_77979785 | 0.67 |
ENSRNOT00000084038
|
AC132740.1
|
|
| chr9_-_44237117 | 0.67 |
ENSRNOT00000068496
|
RGD1310819
|
similar to putative protein (5S487) |
| chr6_+_127941526 | 0.61 |
ENSRNOT00000033897
|
LOC500712
|
Ab1-233 |
| chr9_-_94634109 | 0.59 |
ENSRNOT00000081131
|
AABR07068274.1
|
|
| chr18_-_15284077 | 0.59 |
ENSRNOT00000046326
|
LOC498826
|
LRRGT00165 |
| chr17_-_86657473 | 0.46 |
ENSRNOT00000078827
|
AABR07028795.1
|
|
| chr8_-_107602263 | 0.45 |
ENSRNOT00000017658
|
Esyt3
|
extended synaptotagmin 3 |
| chrX_+_62366453 | 0.43 |
ENSRNOT00000089828
|
Arx
|
aristaless related homeobox |
| chr3_-_2719513 | 0.41 |
ENSRNOT00000020997
|
Lcn12
|
lipocalin 12 |
| chr14_-_6369666 | 0.41 |
ENSRNOT00000093293
ENSRNOT00000093262 |
Zfp951
|
zinc finger protein 951 |
| chr1_+_267618565 | 0.39 |
ENSRNOT00000076251
|
Gsto2
|
glutathione S-transferase omega 2 |
| chr4_+_88834066 | 0.35 |
ENSRNOT00000009546
|
Abcg2
|
ATP-binding cassette, subfamily G (WHITE), member 2 |
| chr12_-_22126350 | 0.35 |
ENSRNOT00000076328
|
Sap25
|
Sin3A-associated protein 25 |
| chr16_-_81714346 | 0.34 |
ENSRNOT00000092552
|
Lamp1
|
lysosomal-associated membrane protein 1 |
| chr17_-_79676499 | 0.33 |
ENSRNOT00000022711
|
Itga8
|
integrin subunit alpha 8 |
| chr5_-_104980641 | 0.33 |
ENSRNOT00000071192
|
Haus6
|
HAUS augmin-like complex, subunit 6 |
| chr1_+_262905570 | 0.32 |
ENSRNOT00000090765
|
Kcnip2
|
potassium voltage-gated channel interacting protein 2 |
| chr1_-_265560386 | 0.32 |
ENSRNOT00000048592
|
LOC100911951
|
Kv channel-interacting protein 2-like |
| chr10_+_64737022 | 0.32 |
ENSRNOT00000017071
ENSRNOT00000093232 ENSRNOT00000017042 ENSRNOT00000093244 |
Lgals9
|
galectin 9 |
| chr1_+_151439409 | 0.31 |
ENSRNOT00000022060
ENSRNOT00000050639 |
Grm5
|
glutamate metabotropic receptor 5 |
| chr6_+_135313008 | 0.30 |
ENSRNOT00000030864
|
Tecpr2
|
tectonin beta-propeller repeat containing 2 |
| chr2_-_230273709 | 0.30 |
ENSRNOT00000012587
|
Mcub
|
mitochondrial calcium uniporter dominant negative beta subunit |
| chr4_-_32392007 | 0.29 |
ENSRNOT00000014946
|
Dlx5
|
distal-less homeobox 5 |
| chr2_-_210943620 | 0.29 |
ENSRNOT00000026750
|
Gpr61
|
G protein-coupled receptor 61 |
| chr7_-_80457816 | 0.28 |
ENSRNOT00000039430
|
AABR07057617.1
|
|
| chr6_-_47848026 | 0.28 |
ENSRNOT00000011048
ENSRNOT00000090017 |
Allc
|
allantoicase |
| chr10_+_45659143 | 0.27 |
ENSRNOT00000058327
|
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr1_+_100299626 | 0.27 |
ENSRNOT00000092327
ENSRNOT00000044257 |
Shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr7_+_143707237 | 0.27 |
ENSRNOT00000074212
|
Tns2
|
tensin 2 |
| chr17_-_76188979 | 0.26 |
ENSRNOT00000092442
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr9_-_49448168 | 0.26 |
ENSRNOT00000059478
|
AABR07067499.1
|
|
| chr20_+_14578605 | 0.26 |
ENSRNOT00000041165
|
Rtdr1
|
rhabdoid tumor deletion region gene 1 |
| chr2_+_248276709 | 0.25 |
ENSRNOT00000068683
|
Gbp2
|
guanylate binding protein 2 |
| chr1_+_99505677 | 0.25 |
ENSRNOT00000024645
|
Zfp719
|
zinc finger protein 719 |
| chr1_-_37818879 | 0.25 |
ENSRNOT00000043747
|
LOC680200
|
similar to zinc finger protein 455 |
| chr8_+_61659445 | 0.25 |
ENSRNOT00000023831
|
Ptpn9
|
protein tyrosine phosphatase, non-receptor type 9 |
| chr12_-_17322608 | 0.24 |
ENSRNOT00000033038
|
LOC102546864
|
uncharacterized LOC102546864 |
| chr15_+_1054937 | 0.24 |
ENSRNOT00000008154
|
AABR07016841.1
|
|
| chr14_-_78377825 | 0.24 |
ENSRNOT00000068104
|
AABR07015812.1
|
|
| chr9_-_79898912 | 0.23 |
ENSRNOT00000022076
|
March4
|
membrane associated ring-CH-type finger 4 |
| chr4_+_62299044 | 0.23 |
ENSRNOT00000032077
|
Agbl3
|
ATP/GTP binding protein-like 3 |
| chr1_+_98627372 | 0.23 |
ENSRNOT00000030370
|
Dzf17
|
zinc finger protein 17 |
| chr1_+_80777014 | 0.23 |
ENSRNOT00000079758
|
Gm19345
|
predicted gene, 19345 |
| chr2_-_219628997 | 0.23 |
ENSRNOT00000064484
|
Trmt13
|
tRNA methyltransferase 13 homolog |
| chr20_+_3189473 | 0.23 |
ENSRNOT00000047439
|
RT1-T24-4
|
RT1 class I, locus T24, gene 4 |
| chr5_-_20189721 | 0.22 |
ENSRNOT00000014661
|
Tox
|
thymocyte selection-associated high mobility group box |
| chr9_+_94279155 | 0.22 |
ENSRNOT00000065805
|
Prss56
|
protease, serine, 56 |
| chr5_+_27312928 | 0.22 |
ENSRNOT00000078102
|
Runx1t1
|
RUNX1 translocation partner 1 |
| chr15_-_34400449 | 0.22 |
ENSRNOT00000048455
|
Rabggta
|
Rab geranylgeranyltransferase, alpha subunit |
| chr17_+_45175121 | 0.22 |
ENSRNOT00000080417
|
Nkapl
|
NFKB activating protein-like |
| chr1_-_164659992 | 0.21 |
ENSRNOT00000024281
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr8_+_61532465 | 0.21 |
ENSRNOT00000023326
|
Cspg4
|
chondroitin sulfate proteoglycan 4 |
| chr16_-_49318383 | 0.21 |
ENSRNOT00000045721
|
Cfap97
|
cilia and flagella associated protein 97 |
| chr4_-_65818521 | 0.21 |
ENSRNOT00000064201
|
Atp6v0a4
|
ATPase H+ transporting V0 subunit a4 |
| chr3_+_148438939 | 0.21 |
ENSRNOT00000064196
|
Ttll9
|
tubulin tyrosine ligase like 9 |
| chr1_+_144601410 | 0.21 |
ENSRNOT00000047408
|
Efl1
|
elongation factor like GTPase 1 |
| chr5_+_169521179 | 0.21 |
ENSRNOT00000067892
|
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr2_+_54191538 | 0.21 |
ENSRNOT00000019524
|
Plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr4_-_159079003 | 0.21 |
ENSRNOT00000026691
|
Kcna5
|
potassium voltage-gated channel subfamily A member 5 |
| chr7_+_18668692 | 0.21 |
ENSRNOT00000009532
|
Kank3
|
KN motif and ankyrin repeat domains 3 |
| chr18_-_70924708 | 0.20 |
ENSRNOT00000025257
|
Lipg
|
lipase G, endothelial type |
| chr12_-_22245100 | 0.20 |
ENSRNOT00000001912
|
Gigyf1
|
GRB10 interacting GYF protein 1 |
| chr10_-_109278510 | 0.20 |
ENSRNOT00000038993
|
Tepsin
|
TEPSIN, adaptor related protein complex 4 accessory protein |
| chr2_+_251534535 | 0.20 |
ENSRNOT00000080311
|
AABR07013701.1
|
|
| chr3_+_2182957 | 0.19 |
ENSRNOT00000011633
|
Pnpla7
|
patatin-like phospholipase domain containing 7 |
| chr4_-_57823283 | 0.19 |
ENSRNOT00000032772
ENSRNOT00000091255 |
Tmem209
|
transmembrane protein 209 |
| chr9_+_12420368 | 0.19 |
ENSRNOT00000071620
|
AABR07066693.1
|
|
| chr10_-_73629581 | 0.19 |
ENSRNOT00000091172
|
Brip1
|
BRCA1 interacting protein C-terminal helicase 1 |
| chr17_+_10463303 | 0.19 |
ENSRNOT00000060822
|
Rnf44
|
ring finger protein 44 |
| chr9_+_66889028 | 0.18 |
ENSRNOT00000087194
|
Carf
|
calcium responsive transcription factor |
| chr11_+_68105369 | 0.18 |
ENSRNOT00000046888
|
Parp14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr8_+_118333706 | 0.18 |
ENSRNOT00000028278
|
Cspg5
|
chondroitin sulfate proteoglycan 5 |
| chr6_+_76349362 | 0.18 |
ENSRNOT00000043224
|
Aldoart2
|
aldolase 1 A retrogene 2 |
| chrX_+_1311121 | 0.18 |
ENSRNOT00000038909
|
Cfp
|
complement factor properdin |
| chr8_+_82038967 | 0.18 |
ENSRNOT00000079535
|
Myo5a
|
myosin VA |
| chr19_-_10653800 | 0.18 |
ENSRNOT00000022128
|
Cx3cl1
|
C-X3-C motif chemokine ligand 1 |
| chr17_-_4454701 | 0.18 |
ENSRNOT00000080750
ENSRNOT00000066950 |
Dapk1
|
death associated protein kinase 1 |
| chr6_-_23543172 | 0.18 |
ENSRNOT00000006073
|
Spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr3_+_152143811 | 0.18 |
ENSRNOT00000026578
|
LOC100911109
|
sperm-associated antigen 4 protein-like |
| chr13_+_70157522 | 0.18 |
ENSRNOT00000036906
|
Apobec4
|
apolipoprotein B mRNA editing enzyme catalytic polypeptide like 4 |
| chr10_+_35857797 | 0.17 |
ENSRNOT00000004517
|
Cby3
|
chibby family member 3 |
| chr12_+_19680712 | 0.17 |
ENSRNOT00000081310
|
AABR07035561.2
|
|
| chr5_-_105579959 | 0.17 |
ENSRNOT00000010827
|
Slc24a2
|
solute carrier family 24 member 2 |
| chr7_-_117267803 | 0.17 |
ENSRNOT00000082271
|
Plec
|
plectin |
| chr14_+_37435654 | 0.17 |
ENSRNOT00000002991
|
Ociad2
|
OCIA domain containing 2 |
| chr1_+_191344979 | 0.17 |
ENSRNOT00000023773
|
Hs3st2
|
heparan sulfate-glucosamine 3-sulfotransferase 2 |
| chr16_+_24797124 | 0.17 |
ENSRNOT00000018976
|
Npy5r
|
neuropeptide Y receptor Y5 |
| chr2_+_69415057 | 0.17 |
ENSRNOT00000013152
|
Cdh10
|
cadherin 10 |
| chr13_-_100740728 | 0.16 |
ENSRNOT00000000074
|
Fbxo28
|
F-box protein 28 |
| chr2_+_187447501 | 0.16 |
ENSRNOT00000038589
|
Iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr13_-_69385074 | 0.16 |
ENSRNOT00000059807
|
RGD1309104
|
similar to RIKEN cDNA 1700025G04 gene |
| chr15_+_18399733 | 0.16 |
ENSRNOT00000061158
|
Fam107a
|
family with sequence similarity 107, member A |
| chr10_-_25910298 | 0.16 |
ENSRNOT00000065633
ENSRNOT00000079646 |
Ccng1
|
cyclin G1 |
| chr10_-_62699723 | 0.16 |
ENSRNOT00000086706
|
Coro6
|
coronin 6 |
| chr7_-_68549763 | 0.16 |
ENSRNOT00000078014
|
Slc16a7
|
solute carrier family 16 member 7 |
| chr7_+_2827247 | 0.15 |
ENSRNOT00000090689
|
Rnf41
|
ring finger protein 41 |
| chr7_-_60860990 | 0.15 |
ENSRNOT00000009511
|
Rap1b
|
RAP1B, member of RAS oncogene family |
| chr15_+_18399515 | 0.15 |
ENSRNOT00000071273
|
Fam107a
|
family with sequence similarity 107, member A |
| chr7_-_130128589 | 0.15 |
ENSRNOT00000079777
ENSRNOT00000009325 |
Mapk11
|
mitogen-activated protein kinase 11 |
| chr3_+_11207542 | 0.15 |
ENSRNOT00000013546
|
Prrc2b
|
proline-rich coiled-coil 2B |
| chr20_+_34633157 | 0.15 |
ENSRNOT00000000469
|
Pln
|
phospholamban |
| chr4_-_108717309 | 0.15 |
ENSRNOT00000085062
|
AABR07061178.1
|
|
| chr2_-_243475639 | 0.15 |
ENSRNOT00000089222
|
RGD1309170
|
similar to hypothetical protein DKFZp434G072 |
| chr5_+_35991068 | 0.15 |
ENSRNOT00000061139
|
Pnisr
|
PNN interacting serine and arginine rich protein |
| chr9_+_46997798 | 0.15 |
ENSRNOT00000087112
ENSRNOT00000082408 |
Il1r1
|
interleukin 1 receptor type 1 |
| chr10_+_46314639 | 0.15 |
ENSRNOT00000089724
|
Med9
|
mediator complex subunit 9 |
| chr6_-_91250138 | 0.14 |
ENSRNOT00000052408
|
LOC100911256
|
ninein-like |
| chr2_+_211176556 | 0.14 |
ENSRNOT00000055880
|
Psrc1
|
proline and serine rich coiled-coil 1 |
| chr10_-_107376645 | 0.14 |
ENSRNOT00000046213
|
Cep295nl
|
CEP295 N-terminal like |
| chr11_+_60102121 | 0.14 |
ENSRNOT00000045521
|
Tmprss7
|
transmembrane protease, serine 7 |
| chr7_-_98098268 | 0.14 |
ENSRNOT00000010361
|
Fbxo32
|
F-box protein 32 |
| chr6_+_33517769 | 0.14 |
ENSRNOT00000007991
|
Hs1bp3
|
HCLS1 binding protein 3 |
| chr11_+_77644541 | 0.14 |
ENSRNOT00000074688
|
Tmem207
|
transmembrane protein 207 |
| chr16_+_61130755 | 0.14 |
ENSRNOT00000042609
|
AABR07026048.1
|
|
| chr6_+_99625306 | 0.14 |
ENSRNOT00000008573
|
Plekhg3
|
pleckstrin homology and RhoGEF domain containing G3 |
| chr3_+_2262253 | 0.14 |
ENSRNOT00000042100
ENSRNOT00000061303 ENSRNOT00000048137 ENSRNOT00000048353 ENSRNOT00000012129 |
Nsmf
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr3_-_94419048 | 0.14 |
ENSRNOT00000015775
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr2_+_104020955 | 0.14 |
ENSRNOT00000045586
|
Mtfr1
|
mitochondrial fission regulator 1 |
| chr7_+_64672722 | 0.14 |
ENSRNOT00000064448
ENSRNOT00000005539 |
Grip1
|
glutamate receptor interacting protein 1 |
| chr5_-_117612123 | 0.14 |
ENSRNOT00000065112
|
Dock7
|
dedicator of cytokinesis 7 |
| chr8_+_70952203 | 0.13 |
ENSRNOT00000019671
|
Mtfmt
|
mitochondrial methionyl-tRNA formyltransferase |
| chr11_-_57993548 | 0.13 |
ENSRNOT00000002957
|
Nectin3
|
nectin cell adhesion molecule 3 |
| chr3_-_51054378 | 0.13 |
ENSRNOT00000089243
|
Grb14
|
growth factor receptor bound protein 14 |
| chr15_-_34694180 | 0.13 |
ENSRNOT00000079505
ENSRNOT00000027950 ENSRNOT00000079782 ENSRNOT00000080221 |
Mcpt8
|
mast cell protease 8 |
| chr1_+_213870502 | 0.13 |
ENSRNOT00000086483
|
B4galnt4
|
beta-1,4-N-acetyl-galactosaminyl transferase 4 |
| chr13_+_83073866 | 0.13 |
ENSRNOT00000075996
|
Dpt
|
dermatopontin |
| chr12_-_38504774 | 0.13 |
ENSRNOT00000011286
|
B3gnt4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr2_-_189254628 | 0.13 |
ENSRNOT00000028234
|
Il6r
|
interleukin 6 receptor |
| chr5_+_124442293 | 0.13 |
ENSRNOT00000041922
|
RGD1564074
|
similar to novel protein |
| chr12_+_45319501 | 0.13 |
ENSRNOT00000090630
ENSRNOT00000041732 |
RGD1561114
|
similar to hypothetical protein 4930474N05 |
| chr8_-_122987191 | 0.13 |
ENSRNOT00000033976
|
Gpd1l
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr5_-_173653905 | 0.13 |
ENSRNOT00000038556
|
Plekhn1
|
pleckstrin homology domain containing N1 |
| chr1_+_87938042 | 0.13 |
ENSRNOT00000027837
|
Map4k1
|
mitogen activated protein kinase kinase kinase kinase 1 |
| chr1_-_100969560 | 0.13 |
ENSRNOT00000035908
|
Cpt1c
|
carnitine palmitoyltransferase 1c |
| chr12_-_3924415 | 0.12 |
ENSRNOT00000067752
|
AABR07035074.1
|
|
| chr4_-_178168690 | 0.12 |
ENSRNOT00000020729
|
Sox5
|
SRY box 5 |
| chr2_+_251002213 | 0.12 |
ENSRNOT00000080600
|
Odf2l
|
outer dense fiber of sperm tails 2-like |
| chr4_-_6062641 | 0.12 |
ENSRNOT00000074846
|
Cct8l1
|
chaperonin containing TCP1, subunit 8 (theta)-like 1 |
| chr8_+_113105814 | 0.12 |
ENSRNOT00000016749
|
Cpne4
|
copine 4 |
| chr10_+_4953879 | 0.12 |
ENSRNOT00000003455
|
Tnp2
|
transition protein 2 |
| chr4_+_157538303 | 0.11 |
ENSRNOT00000086418
|
Zfp384
|
zinc finger protein 384 |
| chr3_+_103753238 | 0.11 |
ENSRNOT00000007144
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr3_+_151609602 | 0.11 |
ENSRNOT00000065052
|
Spag4
|
sperm associated antigen 4 |
| chr10_+_56381813 | 0.11 |
ENSRNOT00000019687
|
Zbtb4
|
zinc finger and BTB domain containing 4 |
| chr2_+_206454208 | 0.11 |
ENSRNOT00000026863
|
Phtf1
|
putative homeodomain transcription factor 1 |
| chr13_-_88536728 | 0.11 |
ENSRNOT00000003950
|
Uhmk1
|
U2AF homology motif kinase 1 |
| chr13_+_109646455 | 0.11 |
ENSRNOT00000073985
|
LOC498316
|
hypothetical LOC498316 |
| chr8_-_55144087 | 0.11 |
ENSRNOT00000039045
|
Dixdc1
|
DIX domain containing 1 |
| chr17_-_27602934 | 0.11 |
ENSRNOT00000079298
|
Rreb1
|
ras responsive element binding protein 1 |
| chr3_+_14304591 | 0.11 |
ENSRNOT00000007776
|
Cntrl
|
centriolin |
| chr14_+_82350734 | 0.10 |
ENSRNOT00000023259
|
Tmem129
|
transmembrane protein 129 |
| chr6_+_48866601 | 0.10 |
ENSRNOT00000077321
ENSRNOT00000079891 |
Pxdn
|
peroxidasin |
| chr8_-_94920441 | 0.10 |
ENSRNOT00000014165
|
Cep162
|
centrosomal protein 162 |
| chr7_+_27081667 | 0.10 |
ENSRNOT00000066143
|
Nfyb
|
nuclear transcription factor Y subunit beta |
| chr3_+_8534440 | 0.10 |
ENSRNOT00000045827
ENSRNOT00000082672 |
Sptan1
|
spectrin, alpha, non-erythrocytic 1 |
| chr5_+_173447784 | 0.10 |
ENSRNOT00000027251
|
Tnfrsf4
|
TNF receptor superfamily member 4 |
| chr10_+_11100917 | 0.10 |
ENSRNOT00000006067
|
Coro7
|
coronin 7 |
| chr7_-_138483612 | 0.10 |
ENSRNOT00000085620
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr4_-_176606382 | 0.10 |
ENSRNOT00000065576
|
Recql
|
RecQ like helicase |
| chr1_-_142615673 | 0.10 |
ENSRNOT00000018021
|
Iqgap1
|
IQ motif containing GTPase activating protein 1 |
| chr1_-_72377434 | 0.10 |
ENSRNOT00000022193
|
Zfp524
|
zinc finger protein 524 |
| chr14_-_106864892 | 0.10 |
ENSRNOT00000090664
|
Otx1
|
orthodenticle homeobox 1 |
| chr4_+_155653718 | 0.10 |
ENSRNOT00000065419
|
Foxj2
|
forkhead box J2 |
| chr1_-_112947399 | 0.10 |
ENSRNOT00000093306
ENSRNOT00000093259 |
Gabra5
|
gamma-aminobutyric acid type A receptor alpha 5 subunit |
| chr3_-_80873887 | 0.10 |
ENSRNOT00000024280
|
Dgkz
|
diacylglycerol kinase zeta |
| chr3_+_92403582 | 0.10 |
ENSRNOT00000064282
|
Pamr1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr2_-_111065413 | 0.10 |
ENSRNOT00000041111
|
Nlgn1
|
neuroligin 1 |
| chr2_-_127754648 | 0.10 |
ENSRNOT00000087535
|
Mfsd8
|
major facilitator superfamily domain containing 8 |
| chr15_-_34693034 | 0.10 |
ENSRNOT00000083314
|
Mcpt8
|
mast cell protease 8 |
| chr7_+_12941994 | 0.10 |
ENSRNOT00000010832
|
Odf3l2
|
outer dense fiber of sperm tails 3-like 2 |
| chr19_-_22194740 | 0.09 |
ENSRNOT00000086187
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr6_-_11686682 | 0.09 |
ENSRNOT00000021998
|
Fbxo11
|
F-box protein 11 |
| chr6_-_26624092 | 0.09 |
ENSRNOT00000008113
|
Trim54
|
tripartite motif-containing 54 |
| chr5_+_63050758 | 0.09 |
ENSRNOT00000009452
|
Tgfbr1
|
transforming growth factor, beta receptor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ar
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0032197 | transposition, RNA-mediated(GO:0032197) |
| 0.2 | 0.7 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.1 | 0.3 | GO:1904612 | glucuronoside metabolic process(GO:0019389) response to 2,3,7,8-tetrachlorodibenzodioxine(GO:1904612) |
| 0.1 | 0.3 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.1 | 0.3 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.3 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.1 | 0.3 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.1 | 0.3 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.1 | 0.4 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.3 | GO:0050893 | sensory processing(GO:0050893) |
| 0.1 | 0.2 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.1 | 0.2 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.0 | 0.1 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.2 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.0 | 0.2 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.0 | 0.4 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.2 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:1905071 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.0 | 0.2 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.2 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.2 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.2 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0042496 | detection of diacyl bacterial lipopeptide(GO:0042496) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.3 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.2 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.1 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.1 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.0 | 0.1 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 0.1 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.2 | GO:0002865 | negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) |
| 0.0 | 0.1 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.1 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 0.0 | 0.1 | GO:1902683 | regulation of organelle transport along microtubule(GO:1902513) regulation of receptor localization to synapse(GO:1902683) negative regulation of synaptic vesicle transport(GO:1902804) |
| 0.0 | 0.1 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 | 0.1 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.1 | GO:1902037 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 | 0.1 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.2 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.1 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.0 | 0.1 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.2 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.2 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.2 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.0 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.3 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.4 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.1 | GO:0051461 | positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.0 | GO:0046967 | cytosol to ER transport(GO:0046967) peptide antigen transport(GO:0046968) |
| 0.0 | 0.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:0042262 | dGTP catabolic process(GO:0006203) DNA protection(GO:0042262) |
| 0.0 | 0.1 | GO:0072347 | response to anesthetic(GO:0072347) |
| 0.0 | 0.1 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.0 | GO:0042531 | regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.1 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.0 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.2 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.0 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.1 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.2 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.0 | GO:0033986 | response to methanol(GO:0033986) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.1 | 0.2 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.1 | 0.3 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 0.2 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.1 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0090534 | calcium ion-transporting ATPase complex(GO:0090534) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.4 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.1 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.3 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.0 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0015292 | uniporter activity(GO:0015292) |
| 0.1 | 0.4 | GO:0016672 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.1 | 0.3 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 0.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.3 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.1 | 0.2 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.1 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.2 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.3 | GO:0071532 | GKAP/Homer scaffold activity(GO:0030160) ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) diacyl lipopeptide binding(GO:0042498) |
| 0.0 | 0.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) steroid sulfotransferase activity(GO:0050294) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.1 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.0 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.0 | 0.0 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.1 | ST STAT3 PATHWAY | STAT3 Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |