Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Alx4
Z-value: 0.37
Transcription factors associated with Alx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Alx4
|
ENSRNOG00000000008 | ALX homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Alx4 | rn6_v1_chr3_+_82548959_82548959 | -0.26 | 6.7e-01 | Click! |
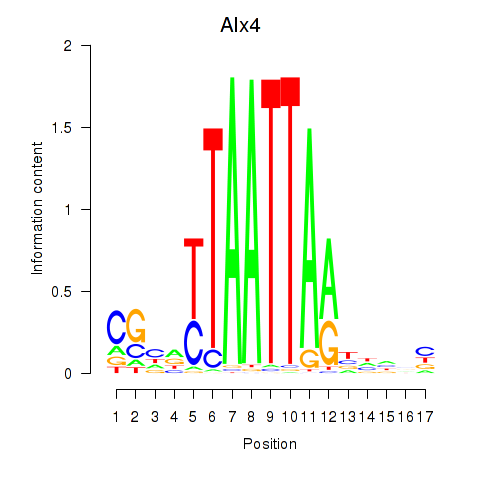
Activity profile of Alx4 motif
Sorted Z-values of Alx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_88670430 | 0.79 |
ENSRNOT00000025547
|
Hcrt
|
hypocretin neuropeptide precursor |
| chr4_+_148782479 | 0.37 |
ENSRNOT00000018133
|
LOC500300
|
similar to hypothetical protein MGC6835 |
| chr12_-_2174131 | 0.30 |
ENSRNOT00000001313
|
Pcp2
|
Purkinje cell protein 2 |
| chr7_-_28711761 | 0.28 |
ENSRNOT00000006249
|
Parpbp
|
PARP1 binding protein |
| chr15_+_37790141 | 0.26 |
ENSRNOT00000076392
ENSRNOT00000091953 |
Il17d
|
interleukin 17D |
| chr2_+_58724855 | 0.25 |
ENSRNOT00000089609
|
Capsl
|
calcyphosine-like |
| chr17_-_61332391 | 0.23 |
ENSRNOT00000034599
|
LOC100362965
|
SNRPN upstream reading frame protein-like |
| chr2_+_145174876 | 0.20 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr5_-_17061361 | 0.19 |
ENSRNOT00000089318
|
Penk
|
proenkephalin |
| chr6_+_8886591 | 0.16 |
ENSRNOT00000091510
ENSRNOT00000089174 |
Six3
|
SIX homeobox 3 |
| chr8_+_22559098 | 0.16 |
ENSRNOT00000041091
|
LOC691141
|
hypothetical protein LOC691141 |
| chr5_+_50381244 | 0.15 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr5_-_17061837 | 0.14 |
ENSRNOT00000011892
|
Penk
|
proenkephalin |
| chr5_-_134927235 | 0.14 |
ENSRNOT00000016751
|
Uqcrh
|
ubiquinol-cytochrome c reductase hinge protein |
| chr2_+_158097843 | 0.14 |
ENSRNOT00000016541
|
Ptx3
|
pentraxin 3 |
| chr3_+_159368273 | 0.14 |
ENSRNOT00000041688
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chrX_+_144994139 | 0.13 |
ENSRNOT00000071783
|
LOC102546596
|
pre-mRNA-splicing factor CWC22 homolog |
| chr10_+_69412017 | 0.13 |
ENSRNOT00000009448
|
Ccl2
|
C-C motif chemokine ligand 2 |
| chr11_+_36851038 | 0.12 |
ENSRNOT00000002221
ENSRNOT00000061047 |
Pcp4
|
Purkinje cell protein 4 |
| chr10_+_43601689 | 0.12 |
ENSRNOT00000029238
|
Mrpl22
|
mitochondrial ribosomal protein L22 |
| chr10_-_91047177 | 0.11 |
ENSRNOT00000003986
|
C1ql1
|
complement C1q like 1 |
| chr13_-_50514151 | 0.11 |
ENSRNOT00000003951
|
Ren
|
renin |
| chr14_-_115052450 | 0.10 |
ENSRNOT00000067998
|
Acyp2
|
acylphosphatase 2 |
| chr20_+_44680449 | 0.10 |
ENSRNOT00000000728
|
Traf3ip2
|
Traf3 interacting protein 2 |
| chr15_+_108318664 | 0.10 |
ENSRNOT00000057469
|
Ubac2
|
UBA domain containing 2 |
| chr12_+_50407843 | 0.10 |
ENSRNOT00000073763
|
Cryba4
|
crystallin, beta A4 |
| chr6_-_104290579 | 0.09 |
ENSRNOT00000066014
|
Erh
|
enhancer of rudimentary homolog (Drosophila) |
| chr4_+_169147243 | 0.09 |
ENSRNOT00000011580
|
Emp1
|
epithelial membrane protein 1 |
| chr10_+_64174931 | 0.09 |
ENSRNOT00000035948
|
RGD1565611
|
RGD1565611 |
| chr3_-_102826379 | 0.09 |
ENSRNOT00000073423
|
Olr769
|
olfactory receptor 769 |
| chr4_-_157304653 | 0.09 |
ENSRNOT00000051613
|
Lrrc23
|
leucine rich repeat containing 23 |
| chr9_+_20241062 | 0.08 |
ENSRNOT00000071593
|
LOC100911585
|
leucine-rich repeat-containing protein 23-like |
| chr14_+_5928737 | 0.08 |
ENSRNOT00000071877
ENSRNOT00000040985 ENSRNOT00000074889 |
Mpa2l
|
macrophage activation 2 like |
| chr1_+_150310319 | 0.08 |
ENSRNOT00000042081
|
Olr34
|
olfactory receptor 34 |
| chr4_+_117962319 | 0.08 |
ENSRNOT00000057441
|
Tgfa
|
transforming growth factor alpha |
| chr20_-_10013190 | 0.08 |
ENSRNOT00000084726
ENSRNOT00000089112 |
Rsph1
|
radial spoke head 1 homolog |
| chr9_+_61692154 | 0.08 |
ENSRNOT00000082300
|
Hspe1
|
heat shock protein family E member 1 |
| chr20_+_4966817 | 0.08 |
ENSRNOT00000081527
ENSRNOT00000081265 |
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr15_-_29369504 | 0.08 |
ENSRNOT00000060297
|
AABR07017624.1
|
|
| chr19_+_26142720 | 0.08 |
ENSRNOT00000005270
|
RGD1564093
|
similar to RIKEN cDNA 2310036O22 |
| chr8_+_71914867 | 0.08 |
ENSRNOT00000023372
|
Dapk2
|
death-associated protein kinase 2 |
| chr11_+_36555416 | 0.07 |
ENSRNOT00000064981
|
Sh3bgr
|
SH3 domain binding glutamate-rich protein |
| chr20_+_42966140 | 0.07 |
ENSRNOT00000000707
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr10_-_29026002 | 0.07 |
ENSRNOT00000005070
|
Pttg1
|
pituitary tumor-transforming 1 |
| chr4_+_88328061 | 0.07 |
ENSRNOT00000084775
|
Vom1r87
|
vomeronasal 1 receptor 87 |
| chr8_+_117117430 | 0.07 |
ENSRNOT00000073247
|
Gpx1
|
glutathione peroxidase 1 |
| chr10_-_105668593 | 0.07 |
ENSRNOT00000016622
|
St6galnac2
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 |
| chr10_-_34961608 | 0.07 |
ENSRNOT00000033056
|
LOC103689931
|
heterogeneous nuclear ribonucleoprotein A/B |
| chrX_-_14972675 | 0.07 |
ENSRNOT00000079664
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr2_+_88217188 | 0.07 |
ENSRNOT00000014267
|
Car1
|
carbonic anhydrase I |
| chr11_+_46184871 | 0.07 |
ENSRNOT00000048417
|
Tfg
|
Trk-fused gene |
| chrX_-_77559348 | 0.07 |
ENSRNOT00000047823
|
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chr11_-_87403367 | 0.07 |
ENSRNOT00000057751
|
Thap7
|
THAP domain containing 7 |
| chr2_+_187447501 | 0.07 |
ENSRNOT00000038589
|
Iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr2_-_185852759 | 0.06 |
ENSRNOT00000049461
|
Mab21l2
|
mab-21 like 2 |
| chrM_+_7758 | 0.06 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr19_-_37938857 | 0.06 |
ENSRNOT00000026730
|
Slc12a4
|
solute carrier family 12 member 4 |
| chr17_-_43776460 | 0.06 |
ENSRNOT00000089055
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chr3_-_82368357 | 0.06 |
ENSRNOT00000000052
|
Cd82
|
Cd82 molecule |
| chr4_+_155321553 | 0.06 |
ENSRNOT00000089614
|
Mfap5
|
microfibrillar associated protein 5 |
| chrX_+_106523278 | 0.06 |
ENSRNOT00000070802
|
MGC109340
|
similar to Microsomal signal peptidase 23 kDa subunit (SPase 22 kDa subunit) (SPC22/23) |
| chr18_+_68408890 | 0.06 |
ENSRNOT00000039702
|
Ccdc68
|
coiled-coil domain containing 68 |
| chr4_+_144382945 | 0.06 |
ENSRNOT00000007601
|
Cav3
|
caveolin 3 |
| chr7_+_2459141 | 0.06 |
ENSRNOT00000075681
|
Naca
|
nascent polypeptide-associated complex alpha subunit |
| chr5_-_38923095 | 0.06 |
ENSRNOT00000009146
|
AABR07047593.1
|
|
| chr6_+_49825469 | 0.06 |
ENSRNOT00000006921
|
Fam150b
|
family with sequence similarity 150, member B |
| chr2_+_188583664 | 0.06 |
ENSRNOT00000045477
|
Dpm3
|
dolichyl-phosphate mannosyltransferase subunit 3 |
| chr6_-_7058314 | 0.06 |
ENSRNOT00000045996
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr7_+_2458833 | 0.06 |
ENSRNOT00000085092
|
Naca
|
nascent polypeptide-associated complex alpha subunit |
| chr6_+_47940183 | 0.06 |
ENSRNOT00000011951
|
Adi1
|
acireductone dioxygenase 1 |
| chr2_+_80269661 | 0.06 |
ENSRNOT00000015975
|
AABR07008940.1
|
|
| chr3_+_7884822 | 0.06 |
ENSRNOT00000019157
|
Med27
|
mediator complex subunit 27 |
| chr4_+_6827429 | 0.06 |
ENSRNOT00000071737
|
Rheb
|
Ras homolog enriched in brain |
| chr14_+_82769642 | 0.06 |
ENSRNOT00000065393
|
Ctbp1
|
C-terminal binding protein 1 |
| chr1_+_213636093 | 0.06 |
ENSRNOT00000019642
|
Psmd13
|
proteasome 26S subunit, non-ATPase 13 |
| chr15_-_95514259 | 0.06 |
ENSRNOT00000038433
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr9_+_100829552 | 0.06 |
ENSRNOT00000024695
|
Bok
|
BOK, BCL2 family apoptosis regulator |
| chr8_+_126975833 | 0.06 |
ENSRNOT00000088348
|
AABR07071701.1
|
|
| chr5_-_22769907 | 0.05 |
ENSRNOT00000047805
ENSRNOT00000076167 ENSRNOT00000076507 ENSRNOT00000076113 ENSRNOT00000083779 |
Asph
|
aspartate-beta-hydroxylase |
| chr10_-_40381886 | 0.05 |
ENSRNOT00000050213
|
LOC100912427
|
nuclease-sensitive element-binding protein 1-like |
| chr3_+_22964230 | 0.05 |
ENSRNOT00000041813
|
LOC100362149
|
ribosomal protein S20-like |
| chr2_+_188784222 | 0.05 |
ENSRNOT00000028095
|
Pmvk
|
phosphomevalonate kinase |
| chr1_-_141504111 | 0.05 |
ENSRNOT00000040164
|
Snrpep2
|
small nuclear ribonucleoprotein polypeptide E pseudogene 2 |
| chr3_-_111087347 | 0.05 |
ENSRNOT00000018277
|
Rhov
|
ras homolog family member V |
| chr1_+_229416489 | 0.05 |
ENSRNOT00000028617
|
Olr336
|
olfactory receptor 336 |
| chrY_-_1305026 | 0.05 |
ENSRNOT00000092901
|
Usp9y
|
ubiquitin specific peptidase 9, Y-linked |
| chr4_+_169161585 | 0.05 |
ENSRNOT00000079785
|
Emp1
|
epithelial membrane protein 1 |
| chr3_-_4341771 | 0.05 |
ENSRNOT00000034694
|
LOC684988
|
similar to ribosomal protein S13 |
| chr10_+_83476107 | 0.05 |
ENSRNOT00000075733
|
Phb
|
prohibitin |
| chr1_-_148443358 | 0.05 |
ENSRNOT00000072973
|
Vbp1
|
VHL binding protein 1 |
| chr5_-_99033107 | 0.05 |
ENSRNOT00000041306
|
Hspa8
|
heat shock protein family A (Hsp70) member 8 |
| chr2_-_123972356 | 0.05 |
ENSRNOT00000023348
|
Il21
|
interleukin 21 |
| chr13_+_56096834 | 0.05 |
ENSRNOT00000035129
|
Dennd1b
|
DENN domain containing 1B |
| chr8_+_100260049 | 0.05 |
ENSRNOT00000011090
|
AABR07071101.1
|
|
| chr1_-_107373807 | 0.05 |
ENSRNOT00000056024
|
Svip
|
small VCP interacting protein |
| chr17_+_44794130 | 0.05 |
ENSRNOT00000077571
|
Hist1h2ac
|
histone cluster 1, H2ac |
| chr3_+_3389612 | 0.05 |
ENSRNOT00000041984
|
Rpl8
|
ribosomal protein L8 |
| chr20_+_19325121 | 0.05 |
ENSRNOT00000058151
|
Phyhipl
|
phytanoyl-CoA 2-hydroxylase interacting protein-like |
| chr9_-_1012450 | 0.05 |
ENSRNOT00000051449
|
LOC100359951
|
ribosomal protein S20-like |
| chr20_+_3176107 | 0.05 |
ENSRNOT00000001036
|
RT1-S3
|
RT1 class Ib, locus S3 |
| chr2_-_147392062 | 0.05 |
ENSRNOT00000021535
|
Tm4sf1
|
transmembrane 4 L six family member 1 |
| chr4_+_29535852 | 0.05 |
ENSRNOT00000087619
|
NEWGENE_621351
|
collagen, type I, alpha 2 |
| chr18_+_70739492 | 0.05 |
ENSRNOT00000085601
|
Acaa2
|
acetyl-CoA acyltransferase 2 |
| chr2_+_93758919 | 0.04 |
ENSRNOT00000077782
|
Fabp12
|
fatty acid binding protein 12 |
| chr2_+_239415046 | 0.04 |
ENSRNOT00000072196
|
Cxxc4
|
CXXC finger protein 4 |
| chr12_+_4310334 | 0.04 |
ENSRNOT00000041244
|
Vom2r60
|
vomeronasal 2 receptor, 60 |
| chr13_-_102857551 | 0.04 |
ENSRNOT00000080309
|
Mark1
|
microtubule affinity regulating kinase 1 |
| chr4_-_115208026 | 0.04 |
ENSRNOT00000084388
ENSRNOT00000066678 |
Dguok
|
deoxyguanosine kinase |
| chr4_+_6946634 | 0.04 |
ENSRNOT00000040373
|
Wdr86
|
WD repeat domain 86 |
| chr1_-_225594958 | 0.04 |
ENSRNOT00000027492
|
Scgb1d2
|
secretoglobin, family 1D, member 2 |
| chr11_-_62067655 | 0.04 |
ENSRNOT00000093382
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chrX_+_29504349 | 0.04 |
ENSRNOT00000005981
|
Tceanc
|
transcription elongation factor A N-terminal and central domain containing |
| chr16_+_2537248 | 0.04 |
ENSRNOT00000017995
|
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr5_+_28485619 | 0.04 |
ENSRNOT00000093341
ENSRNOT00000093129 |
LOC100912373
|
uncharacterized LOC100912373 |
| chr3_+_173799833 | 0.04 |
ENSRNOT00000081235
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr20_-_5020150 | 0.04 |
ENSRNOT00000001146
|
Sapcd1
|
suppressor APC domain containing 1 |
| chr2_+_187951344 | 0.04 |
ENSRNOT00000027123
|
Ssr2
|
signal sequence receptor, beta |
| chr15_-_37830131 | 0.04 |
ENSRNOT00000075861
|
Eef1akmt1
|
eukaryotic translation elongation factor 1 alpha lysine methyltransferase 1 |
| chr10_+_106812739 | 0.04 |
ENSRNOT00000074225
|
Syngr2
|
synaptogyrin 2 |
| chr13_-_80862963 | 0.04 |
ENSRNOT00000004864
|
Fmo3
|
flavin containing monooxygenase 3 |
| chr5_+_145311375 | 0.04 |
ENSRNOT00000019224
|
Smim12
|
small integral membrane protein 12 |
| chr14_-_6900733 | 0.04 |
ENSRNOT00000061224
|
Dmp1
|
dentin matrix acidic phosphoprotein 1 |
| chr20_+_8484311 | 0.04 |
ENSRNOT00000050041
|
LOC100364116
|
ribosomal protein S20-like |
| chr9_+_10941613 | 0.04 |
ENSRNOT00000070794
|
Sema6b
|
semaphorin 6B |
| chr10_-_86393141 | 0.04 |
ENSRNOT00000009485
|
Mien1
|
migration and invasion enhancer 1 |
| chr2_-_170301348 | 0.04 |
ENSRNOT00000088131
|
Si
|
sucrase-isomaltase |
| chrX_-_121731543 | 0.04 |
ENSRNOT00000018788
|
Klhl13
|
kelch-like family member 13 |
| chr2_-_158156444 | 0.04 |
ENSRNOT00000088559
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr1_-_89014189 | 0.04 |
ENSRNOT00000028413
|
Lin37
|
lin-37 DREAM MuvB core complex component |
| chr11_-_17684903 | 0.04 |
ENSRNOT00000051213
|
Tmprss15
|
transmembrane protease, serine 15 |
| chr9_-_95362014 | 0.04 |
ENSRNOT00000051065
|
Hjurp
|
Holliday junction recognition protein |
| chr2_-_143104412 | 0.04 |
ENSRNOT00000058116
|
Ufm1
|
ubiquitin-fold modifier 1 |
| chr16_-_36161089 | 0.04 |
ENSRNOT00000017888
|
Scrg1
|
stimulator of chondrogenesis 1 |
| chr2_+_122877286 | 0.04 |
ENSRNOT00000033080
|
Arse
|
arylsulfatase E |
| chr6_+_56846789 | 0.04 |
ENSRNOT00000032108
|
Agmo
|
alkylglycerol monooxygenase |
| chr16_-_7758189 | 0.04 |
ENSRNOT00000026588
|
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr10_-_83898527 | 0.04 |
ENSRNOT00000009815
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr1_-_189238776 | 0.04 |
ENSRNOT00000020817
|
Pdilt
|
protein disulfide isomerase-like, testis expressed |
| chr13_+_76962504 | 0.04 |
ENSRNOT00000076581
|
Rfwd2
|
ring finger and WD repeat domain 2 |
| chr7_-_143967484 | 0.04 |
ENSRNOT00000081758
|
Sp7
|
Sp7 transcription factor |
| chr3_-_94808861 | 0.04 |
ENSRNOT00000038464
|
Prrg4
|
proline rich and Gla domain 4 |
| chr4_-_77489535 | 0.04 |
ENSRNOT00000008728
|
Pdia4
|
protein disulfide isomerase family A, member 4 |
| chr15_+_33606124 | 0.04 |
ENSRNOT00000065210
|
AC115371.1
|
|
| chr4_-_147719072 | 0.04 |
ENSRNOT00000014493
|
Rpl32
|
ribosomal protein L32 |
| chr4_-_155051429 | 0.04 |
ENSRNOT00000020094
|
Klrg1
|
killer cell lectin like receptor G1 |
| chr8_-_121973125 | 0.03 |
ENSRNOT00000012114
|
Pdcd6ip
|
programmed cell death 6 interacting protein |
| chrX_+_110789269 | 0.03 |
ENSRNOT00000086014
|
Rnf128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr6_-_127319362 | 0.03 |
ENSRNOT00000012256
|
Ddx24
|
DEAD-box helicase 24 |
| chrX_+_112311251 | 0.03 |
ENSRNOT00000086698
|
AABR07040855.1
|
|
| chr1_-_102699442 | 0.03 |
ENSRNOT00000056109
|
Tph1
|
tryptophan hydroxylase 1 |
| chr11_+_64472072 | 0.03 |
ENSRNOT00000042756
|
RGD1563835
|
similar to ribosomal protein L27 |
| chrX_-_63291107 | 0.03 |
ENSRNOT00000092019
|
Eif2s3
|
eukaryotic translation initiation factor 2 subunit gamma |
| chr18_+_55466373 | 0.03 |
ENSRNOT00000074629
|
LOC102555392
|
interferon-inducible GTPase 1-like |
| chr17_-_89163113 | 0.03 |
ENSRNOT00000050445
|
LOC100360843
|
ribosomal protein S19-like |
| chr1_+_60884761 | 0.03 |
ENSRNOT00000088689
|
LOC100365363
|
vomeronasal 1 receptor, F4-like |
| chr3_-_165537940 | 0.03 |
ENSRNOT00000071119
ENSRNOT00000070964 |
Sall4
|
spalt-like transcription factor 4 |
| chr12_+_45026886 | 0.03 |
ENSRNOT00000001500
|
Pebp1
|
phosphatidylethanolamine binding protein 1 |
| chr5_+_165724027 | 0.03 |
ENSRNOT00000018000
|
Casz1
|
castor zinc finger 1 |
| chr1_+_88113445 | 0.03 |
ENSRNOT00000056955
|
Ggn
|
gametogenetin |
| chr1_+_85213652 | 0.03 |
ENSRNOT00000092044
|
Nccrp1
|
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
| chr6_+_101532518 | 0.03 |
ENSRNOT00000075054
|
Gphn
|
gephyrin |
| chr3_-_95007712 | 0.03 |
ENSRNOT00000017409
|
Eif3m
|
eukaryotic translation initiation factor 3, subunit M |
| chr2_-_181900856 | 0.03 |
ENSRNOT00000082156
|
Lrat
|
lecithin-retinol acyltransferase (phosphatidylcholine-retinol-O-acyltransferase) |
| chr2_+_202200797 | 0.03 |
ENSRNOT00000042263
ENSRNOT00000071938 |
Spag17
|
sperm associated antigen 17 |
| chr12_+_1460538 | 0.03 |
ENSRNOT00000001444
|
Rfc3
|
replication factor C subunit 3 |
| chr1_+_23906717 | 0.03 |
ENSRNOT00000022798
|
Tcf21
|
transcription factor 21 |
| chr18_+_55666027 | 0.03 |
ENSRNOT00000045950
|
RGD1305184
|
similar to CDNA sequence BC023105 |
| chr9_+_8054466 | 0.03 |
ENSRNOT00000081513
|
Adgre4
|
adhesion G protein-coupled receptor E4 |
| chr18_-_3676188 | 0.03 |
ENSRNOT00000073811
|
Ankrd29
|
ankyrin repeat domain 29 |
| chr4_-_100252755 | 0.03 |
ENSRNOT00000017301
|
Vamp8
|
vesicle-associated membrane protein 8 |
| chr2_-_102838745 | 0.03 |
ENSRNOT00000082084
|
Cyp7b1
|
cytochrome P450, family 7, subfamily b, polypeptide 1 |
| chr2_+_54466280 | 0.03 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr1_+_185863043 | 0.03 |
ENSRNOT00000079072
|
Sox6
|
SRY box 6 |
| chrX_+_74205842 | 0.03 |
ENSRNOT00000077003
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr4_+_88423412 | 0.03 |
ENSRNOT00000040163
|
Vom1r88
|
vomeronasal 1 receptor 88 |
| chr10_+_3227160 | 0.03 |
ENSRNOT00000088075
|
Ntan1
|
N-terminal asparagine amidase |
| chr10_-_88266210 | 0.03 |
ENSRNOT00000090702
ENSRNOT00000020603 |
Hap1
|
huntingtin-associated protein 1 |
| chr12_+_46042413 | 0.03 |
ENSRNOT00000046882
|
Ccdc60
|
coiled-coil domain containing 60 |
| chr17_+_15814132 | 0.03 |
ENSRNOT00000032997
|
Susd3
|
sushi domain containing 3 |
| chr7_-_49741540 | 0.03 |
ENSRNOT00000006523
|
Myf6
|
myogenic factor 6 |
| chr10_+_31561895 | 0.03 |
ENSRNOT00000048485
|
Havcr2
|
hepatitis A virus cellular receptor 2 |
| chr6_-_125590049 | 0.03 |
ENSRNOT00000006371
|
Tc2n
|
tandem C2 domains, nuclear |
| chr10_+_5199226 | 0.03 |
ENSRNOT00000003544
|
Dexi
|
dexamethasone-induced transcript |
| chrX_-_138118696 | 0.03 |
ENSRNOT00000084130
|
Frmd7
|
FERM domain containing 7 |
| chr16_-_49453394 | 0.03 |
ENSRNOT00000041617
|
Lrp2bp
|
Lrp2 binding protein |
| chr1_-_185569190 | 0.03 |
ENSRNOT00000090773
|
RGD1311703
|
similar to sid2057p |
| chr4_-_146954159 | 0.03 |
ENSRNOT00000010397
|
Tamm41
|
TAM41 mitochondrial translocator assembly and maintenance homolog |
| chr19_+_29320701 | 0.03 |
ENSRNOT00000049600
|
Polr2m
|
polymerase (RNA) II (DNA directed) polypeptide M |
| chr5_+_10178302 | 0.03 |
ENSRNOT00000009679
|
Sntg1
|
syntrophin, gamma 1 |
| chr1_-_80271001 | 0.03 |
ENSRNOT00000034266
|
Cd3eap
|
CD3e molecule associated protein |
| chr16_+_39145230 | 0.03 |
ENSRNOT00000092942
|
Adam21
|
ADAM metallopeptidase domain 21 |
| chr20_-_11721838 | 0.03 |
ENSRNOT00000001636
|
Ube2g2
|
ubiquitin-conjugating enzyme E2G 2 |
| chr5_-_161981441 | 0.03 |
ENSRNOT00000020316
|
Pdpn
|
podoplanin |
| chr2_+_30685840 | 0.03 |
ENSRNOT00000031385
|
Ccdc125
|
coiled-coil domain containing 125 |
| chr1_-_67065797 | 0.03 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chr10_-_34961349 | 0.03 |
ENSRNOT00000004885
|
LOC103689931
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr4_+_8066907 | 0.03 |
ENSRNOT00000080811
|
Srpk2
|
SRSF protein kinase 2 |
| chr6_+_137323713 | 0.03 |
ENSRNOT00000029017
|
Pld4
|
phospholipase D family, member 4 |
| chr8_-_107490093 | 0.03 |
ENSRNOT00000046832
|
LOC684466
|
similar to Fas apoptotic inhibitory molecule 1 (rFAIM) |
| chr14_-_84334066 | 0.03 |
ENSRNOT00000006160
|
Mtfp1
|
mitochondrial fission process 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Alx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.3 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 0.1 | 0.2 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:0052203 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.0 | 0.1 | GO:0002434 | immune complex clearance(GO:0002434) |
| 0.0 | 0.1 | GO:1901227 | negative regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901227) |
| 0.0 | 0.2 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.1 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.0 | 0.1 | GO:1904009 | cellular response to monosodium glutamate(GO:1904009) |
| 0.0 | 0.3 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:2000424 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.0 | 0.1 | GO:0009609 | response to symbiotic bacterium(GO:0009609) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0002018 | angiotensin maturation(GO:0002003) renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.1 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.1 | GO:1904708 | granulosa cell apoptotic process(GO:1904700) regulation of granulosa cell apoptotic process(GO:1904708) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.2 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.0 | GO:0032759 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.0 | 0.0 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.0 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.1 | GO:0031584 | activation of phospholipase D activity(GO:0031584) regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.0 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.0 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.0 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.0 | GO:1903595 | pancreatic amylase secretion(GO:0036395) regulation of pancreatic amylase secretion(GO:1902276) positive regulation of histamine secretion by mast cell(GO:1903595) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0044307 | germinal vesicle(GO:0042585) dendritic branch(GO:0044307) |
| 0.0 | 0.0 | GO:1990836 | lysosomal matrix(GO:1990836) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.0 | GO:0032398 | MHC class Ib protein complex(GO:0032398) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.0 | GO:0035577 | azurophil granule membrane(GO:0035577) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.0 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.0 | GO:0008396 | oxysterol 7-alpha-hydroxylase activity(GO:0008396) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |