Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Aire
Z-value: 0.47
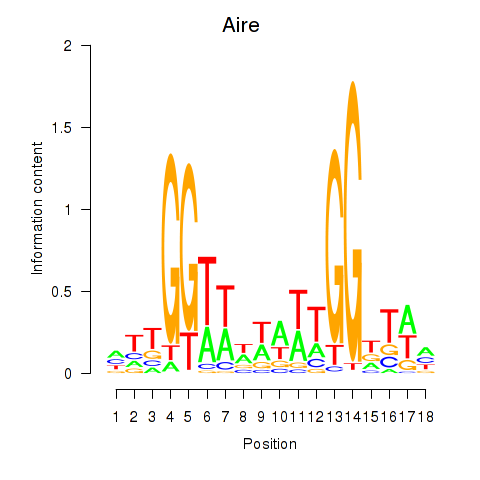
Transcription factors associated with Aire
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Aire
|
ENSRNOG00000001213 | autoimmune regulator |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Aire | rn6_v1_chr20_+_11365697_11365697 | -0.29 | 6.4e-01 | Click! |
Activity profile of Aire motif
Sorted Z-values of Aire motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_23135354 | 0.14 |
ENSRNOT00000011432
|
Steap4
|
STEAP4 metalloreductase |
| chr2_+_209661244 | 0.12 |
ENSRNOT00000091973
|
AABR07012826.1
|
|
| chrM_+_9451 | 0.12 |
ENSRNOT00000041241
|
Mt-nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chrM_+_14136 | 0.11 |
ENSRNOT00000042098
|
Mt-cyb
|
mitochondrially encoded cytochrome b |
| chr8_+_52751854 | 0.09 |
ENSRNOT00000072518
|
Nxpe1
|
neurexophilin and PC-esterase domain family, member 1 |
| chr15_-_83494423 | 0.09 |
ENSRNOT00000037588
|
Dis3
|
DIS3 homolog, exosome endoribonuclease and 3'-5' exoribonuclease |
| chrM_+_7006 | 0.09 |
ENSRNOT00000043693
|
Mt-co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr18_-_36513317 | 0.08 |
ENSRNOT00000025453
|
Plac8l1
|
PLAC8-like 1 |
| chr20_+_34633157 | 0.08 |
ENSRNOT00000000469
|
Pln
|
phospholamban |
| chr1_-_66212418 | 0.08 |
ENSRNOT00000026074
|
LOC691722
|
hypothetical protein LOC691722 |
| chr3_-_161294125 | 0.08 |
ENSRNOT00000021143
|
Spata25
|
spermatogenesis associated 25 |
| chrM_+_11736 | 0.08 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr13_-_82129989 | 0.08 |
ENSRNOT00000078963
|
AC124874.1
|
|
| chrX_+_22285184 | 0.08 |
ENSRNOT00000086262
|
Iqsec2
|
IQ motif and Sec7 domain 2 |
| chr2_+_92573004 | 0.07 |
ENSRNOT00000065774
|
tGap1
|
GTPase activating protein testicular GAP1 |
| chr3_+_161413298 | 0.07 |
ENSRNOT00000023965
ENSRNOT00000088776 |
Mmp9
|
matrix metallopeptidase 9 |
| chr1_+_141120166 | 0.07 |
ENSRNOT00000050759
|
Fanci
|
Fanconi anemia, complementation group I |
| chr6_-_99843245 | 0.07 |
ENSRNOT00000080270
|
Gpx2
|
glutathione peroxidase 2 |
| chr4_-_164123974 | 0.07 |
ENSRNOT00000082447
|
LOC502907
|
similar to immunoreceptor Ly49si1 |
| chrM_+_8599 | 0.07 |
ENSRNOT00000049683
|
Mt-cox3
|
mitochondrially encoded cytochrome C oxidase III |
| chrM_-_14061 | 0.06 |
ENSRNOT00000051268
|
Mt-nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr16_-_79671719 | 0.06 |
ENSRNOT00000015908
|
Myom2
|
myomesin 2 |
| chrX_-_915953 | 0.06 |
ENSRNOT00000075264
|
Spaca5
|
sperm acrosome associated 5 |
| chr14_+_91557601 | 0.06 |
ENSRNOT00000038733
|
RGD1309870
|
hypothetical LOC289778 |
| chr17_+_16455026 | 0.05 |
ENSRNOT00000022932
|
Zfp169
|
zinc finger protein 169 |
| chr1_-_146556171 | 0.05 |
ENSRNOT00000017636
|
Arnt2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chr7_+_70445366 | 0.05 |
ENSRNOT00000045687
|
RGD1565117
|
similar to 40S ribosomal protein S26 |
| chr10_-_34242985 | 0.05 |
ENSRNOT00000046438
|
RGD1559575
|
similar to novel protein |
| chr9_+_12475006 | 0.05 |
ENSRNOT00000079703
|
LOC100912293
|
uncharacterized LOC100912293 |
| chr5_+_122508388 | 0.05 |
ENSRNOT00000038410
|
Tctex1d1
|
Tctex1 domain containing 1 |
| chrX_-_156270748 | 0.05 |
ENSRNOT00000078438
|
Ikbkg
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr2_-_179704629 | 0.05 |
ENSRNOT00000083361
ENSRNOT00000077941 |
Gria2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr19_+_15081590 | 0.05 |
ENSRNOT00000024187
|
Ces1f
|
carboxylesterase 1F |
| chr18_+_61377051 | 0.04 |
ENSRNOT00000066659
|
Oacyl
|
O-acyltransferase like |
| chr15_+_61879184 | 0.04 |
ENSRNOT00000042606
|
Sugt1
|
SGT1 homolog, MIS12 kinetochore complex assembly cochaperone |
| chr7_-_98804499 | 0.04 |
ENSRNOT00000070909
|
Tatdn1
|
TatD DNase domain containing 1 |
| chr8_-_52828134 | 0.04 |
ENSRNOT00000007736
|
Rbm7
|
RNA binding motif protein 7 |
| chr2_-_21931720 | 0.04 |
ENSRNOT00000018449
|
Msh3
|
mutS homolog 3 |
| chr14_-_34561696 | 0.04 |
ENSRNOT00000059763
|
Srd5a3
|
steroid 5 alpha-reductase 3 |
| chr1_-_154165524 | 0.04 |
ENSRNOT00000023468
|
Hikeshi
|
Hikeshi, heat shock protein nuclear import factor |
| chr7_-_11754508 | 0.04 |
ENSRNOT00000026341
|
Oaz1
|
ornithine decarboxylase antizyme 1 |
| chr8_-_23282989 | 0.04 |
ENSRNOT00000019204
|
Zfp717
|
zinc finger protein 717 |
| chr11_+_43521054 | 0.04 |
ENSRNOT00000041679
|
Olr1551
|
olfactory receptor 1551 |
| chr10_-_88611105 | 0.04 |
ENSRNOT00000024718
|
Dhx58
|
DEXH-box helicase 58 |
| chr1_+_101884276 | 0.04 |
ENSRNOT00000082917
|
Tmem143
|
transmembrane protein 143 |
| chr1_+_78739930 | 0.04 |
ENSRNOT00000021976
|
Strn4
|
striatin 4 |
| chr4_+_123162086 | 0.04 |
ENSRNOT00000011557
|
Lsm3
|
LSM3 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr2_+_182006242 | 0.04 |
ENSRNOT00000064091
|
Fga
|
fibrinogen alpha chain |
| chr4_+_88695590 | 0.04 |
ENSRNOT00000087835
|
Ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr5_+_118418799 | 0.04 |
ENSRNOT00000012052
|
Alg6
|
ALG6, alpha-1,3-glucosyltransferase |
| chr20_+_22728208 | 0.04 |
ENSRNOT00000000794
|
LOC100363472
|
nuclear receptor-binding factor 2-like |
| chr16_-_52127591 | 0.03 |
ENSRNOT00000033652
|
Triml2
|
tripartite motif family-like 2 |
| chr4_-_117575154 | 0.03 |
ENSRNOT00000075813
|
LOC102556148
|
probable N-acetyltransferase CML2-like |
| chr13_-_78851600 | 0.03 |
ENSRNOT00000076985
|
Zbtb37
|
zinc finger and BTB domain containing 37 |
| chr11_-_4332255 | 0.03 |
ENSRNOT00000087133
|
Cadm2
|
cell adhesion molecule 2 |
| chr11_+_84745904 | 0.03 |
ENSRNOT00000002617
|
Klhl6
|
kelch-like family member 6 |
| chr1_+_101884019 | 0.03 |
ENSRNOT00000028650
|
Tmem143
|
transmembrane protein 143 |
| chrX_-_29825439 | 0.03 |
ENSRNOT00000048155
|
Gemin8
|
gem (nuclear organelle) associated protein 8 |
| chr16_-_62164135 | 0.03 |
ENSRNOT00000058872
|
Gtf2e2
|
general transcription factor IIE subunit 2 |
| chr4_-_120559078 | 0.03 |
ENSRNOT00000085730
ENSRNOT00000079575 |
Kbtbd12
|
kelch repeat and BTB domain containing 12 |
| chr10_-_52290657 | 0.03 |
ENSRNOT00000005293
|
Map2k4
|
mitogen activated protein kinase kinase 4 |
| chr1_+_228142778 | 0.03 |
ENSRNOT00000028517
|
Mrpl16
|
mitochondrial ribosomal protein L16 |
| chr11_-_31772984 | 0.03 |
ENSRNOT00000002771
|
Dnajc28
|
DnaJ heat shock protein family (Hsp40) member C28 |
| chr13_-_42885440 | 0.03 |
ENSRNOT00000038020
|
Nckap5
|
NCK-associated protein 5 |
| chr3_-_51612397 | 0.03 |
ENSRNOT00000081401
|
Scn3a
|
sodium voltage-gated channel alpha subunit 3 |
| chr1_+_220096422 | 0.03 |
ENSRNOT00000034771
|
Ccdc87
|
coiled-coil domain containing 87 |
| chr15_-_52214616 | 0.03 |
ENSRNOT00000015035
|
Sftpc
|
surfactant protein C |
| chrX_-_45284341 | 0.03 |
ENSRNOT00000045436
|
Obp1f
|
odorant binding protein I f |
| chr8_-_58253688 | 0.02 |
ENSRNOT00000010956
|
Cul5
|
cullin 5 |
| chr8_-_36438730 | 0.02 |
ENSRNOT00000015366
|
Fam118b
|
family with sequence similarity 118, member B |
| chrX_+_83926513 | 0.02 |
ENSRNOT00000035274
|
RGD1561958
|
similar to RIKEN cDNA 2010106E10 |
| chr9_+_84410972 | 0.02 |
ENSRNOT00000019724
|
Mogat1
|
monoacylglycerol O-acyltransferase 1 |
| chr3_-_13978224 | 0.02 |
ENSRNOT00000025528
|
Phf19
|
PHD finger protein 19 |
| chr7_-_123655896 | 0.02 |
ENSRNOT00000012413
|
Cyp2d2
|
cytochrome P450, family 2, subfamily d, polypeptide 2 |
| chr11_+_45462345 | 0.02 |
ENSRNOT00000029420
|
Nit2
|
nitrilase family, member 2 |
| chr4_-_144620731 | 0.02 |
ENSRNOT00000066468
|
Rad18
|
RAD18 E3 ubiquitin protein ligase |
| chr15_-_37830131 | 0.02 |
ENSRNOT00000075861
|
Eef1akmt1
|
eukaryotic translation elongation factor 1 alpha lysine methyltransferase 1 |
| chr7_-_119712888 | 0.02 |
ENSRNOT00000077438
|
Il2rb
|
interleukin 2 receptor subunit beta |
| chr7_-_101138373 | 0.02 |
ENSRNOT00000043257
|
LOC500877
|
Ab1-152 |
| chr9_+_93086012 | 0.02 |
ENSRNOT00000089470
|
Psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr10_-_107158997 | 0.02 |
ENSRNOT00000004047
|
Dnah17
|
dynein, axonemal, heavy chain 17 |
| chr7_-_138483612 | 0.02 |
ENSRNOT00000085620
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr3_+_2872031 | 0.02 |
ENSRNOT00000023062
|
Lcn8
|
lipocalin 8 |
| chr18_-_36322320 | 0.02 |
ENSRNOT00000060260
|
Grxcr2
|
glutaredoxin and cysteine rich domain containing 2 |
| chr9_+_95285592 | 0.02 |
ENSRNOT00000063853
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr11_+_54619129 | 0.02 |
ENSRNOT00000059924
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr17_-_15566332 | 0.02 |
ENSRNOT00000093743
|
Ecm2
|
extracellular matrix protein 2 |
| chr10_-_70735742 | 0.02 |
ENSRNOT00000077035
|
Heatr9
|
HEAT repeat containing 9 |
| chr14_+_79339258 | 0.02 |
ENSRNOT00000058414
|
Tbc1d14
|
TBC1 domain family, member 14 |
| chr14_-_17436897 | 0.02 |
ENSRNOT00000003277
|
Uso1
|
USO1 vesicle transport factor |
| chr13_+_60545635 | 0.02 |
ENSRNOT00000077153
|
Uchl5
|
ubiquitin C-terminal hydrolase L5 |
| chr12_+_4248808 | 0.02 |
ENSRNOT00000042410
|
AABR07035089.1
|
|
| chr1_+_266482858 | 0.01 |
ENSRNOT00000027223
|
As3mt
|
arsenite methyltransferase |
| chr1_+_100059967 | 0.01 |
ENSRNOT00000081095
|
Klk1
|
kallikrein 1 |
| chr2_+_196608496 | 0.01 |
ENSRNOT00000091681
|
Arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr10_-_87153982 | 0.01 |
ENSRNOT00000014414
|
Krt222
|
keratin 222 |
| chr17_+_68477446 | 0.01 |
ENSRNOT00000086236
ENSRNOT00000068174 |
Pitrm1
|
pitrilysin metallopeptidase 1 |
| chr3_-_11820549 | 0.01 |
ENSRNOT00000020660
|
Cfap157
|
cilia and flagella associated protein 157 |
| chr4_-_99546905 | 0.01 |
ENSRNOT00000077447
|
Kdm3a
|
lysine demethylase 3A |
| chr3_-_148493225 | 0.01 |
ENSRNOT00000012141
|
Pdrg1
|
p53 and DNA damage regulated 1 |
| chr6_+_104611026 | 0.01 |
ENSRNOT00000007583
|
Srsf5
|
serine and arginine rich splicing factor 5 |
| chr1_+_59156251 | 0.01 |
ENSRNOT00000017442
|
Lix1
|
limb and CNS expressed 1 |
| chr2_-_100249811 | 0.01 |
ENSRNOT00000086760
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr1_-_162713610 | 0.01 |
ENSRNOT00000018091
|
Aqp11
|
aquaporin 11 |
| chr5_-_124403195 | 0.01 |
ENSRNOT00000067850
|
C8a
|
complement C8 alpha chain |
| chr13_-_98529040 | 0.01 |
ENSRNOT00000091715
|
Psen2
|
presenilin 2 |
| chr1_+_63842277 | 0.01 |
ENSRNOT00000087957
ENSRNOT00000080820 |
Lilrb3a
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3A |
| chr9_+_92618550 | 0.01 |
ENSRNOT00000078987
|
Sp140
|
SP140 nuclear body protein |
| chr18_+_29506128 | 0.01 |
ENSRNOT00000003878
|
Slc35a4
|
solute carrier family 35, member A4 |
| chr8_+_82171497 | 0.01 |
ENSRNOT00000011768
|
Myo5c
|
myosin VC |
| chr1_+_143675985 | 0.01 |
ENSRNOT00000078500
|
Tm6sf1
|
transmembrane 6 superfamily member 1 |
| chr15_+_62520953 | 0.01 |
ENSRNOT00000040851
|
LOC306079
|
similar to RIKEN cDNA 3100001N19 |
| chr3_+_95233874 | 0.01 |
ENSRNOT00000079743
|
LOC691083
|
hypothetical protein LOC691083 |
| chrX_+_26294066 | 0.01 |
ENSRNOT00000037862
|
Hccs
|
holocytochrome c synthase |
| chr3_+_153398130 | 0.01 |
ENSRNOT00000068135
|
Rpn2
|
ribophorin II |
| chr2_-_209001895 | 0.01 |
ENSRNOT00000055960
|
RGD1309110
|
similar to Hypothetical protein MGC58999 |
| chr3_-_153246433 | 0.01 |
ENSRNOT00000067748
|
Samhd1
|
SAM and HD domain containing deoxynucleoside triphosphate triphosphohydrolase 1 |
| chr17_+_25228437 | 0.01 |
ENSRNOT00000072904
|
AABR07027342.1
|
|
| chr7_-_143738237 | 0.01 |
ENSRNOT00000055320
|
Spryd3
|
SPRY domain containing 3 |
| chr5_+_60528997 | 0.01 |
ENSRNOT00000051445
|
Grhpr
|
glyoxylate and hydroxypyruvate reductase |
| chr12_-_19439977 | 0.01 |
ENSRNOT00000060035
|
Nxpe5
|
neurexophilin and PC-esterase domain family, member 5 |
| chr1_-_99135977 | 0.01 |
ENSRNOT00000056515
|
Vom2r38
|
vomeronasal 2 receptor, 38 |
| chr12_+_40417944 | 0.01 |
ENSRNOT00000073125
|
Acad10
|
acyl-CoA dehydrogenase family, member 10 |
| chr4_+_118160147 | 0.01 |
ENSRNOT00000022014
|
Fam136a
|
family with sequence similarity 136, member A |
| chrX_-_1704033 | 0.01 |
ENSRNOT00000051956
|
Usp11
|
ubiquitin specific peptidase 11 |
| chr5_+_15043955 | 0.01 |
ENSRNOT00000047093
|
Rp1
|
retinitis pigmentosa 1 |
| chrX_-_10218583 | 0.00 |
ENSRNOT00000013382
|
Nyx
|
nyctalopin |
| chr9_+_7643533 | 0.00 |
ENSRNOT00000074923
|
Vom2r76
|
vomeronasal 2 receptor, 76 |
| chr11_+_83986230 | 0.00 |
ENSRNOT00000002331
ENSRNOT00000073703 |
Alg3
|
ALG3, alpha-1,3- mannosyltransferase |
| chr8_-_16486419 | 0.00 |
ENSRNOT00000033223
|
LOC690235
|
similar to MAP/microtubule affinity-regulating kinase 4 (MAP/microtubule affinity-regulating kinase-like 1) |
| chr10_+_37594578 | 0.00 |
ENSRNOT00000007676
|
Skp1
|
S-phase kinase-associated protein 1 |
| chr1_+_142951094 | 0.00 |
ENSRNOT00000077441
|
Slc28a1
|
solute carrier family 28 member 1 |
| chr11_+_34101197 | 0.00 |
ENSRNOT00000002299
|
Chaf1b
|
chromatin assembly factor 1 subunit B |
| chr8_-_32000378 | 0.00 |
ENSRNOT00000013341
|
Adamts15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15 |
| chr10_-_56511583 | 0.00 |
ENSRNOT00000021402
|
LOC497940
|
similar to RIKEN cDNA 2810408A11 |
| chr7_+_123482255 | 0.00 |
ENSRNOT00000064487
|
LOC688613
|
hypothetical protein LOC688613 |
| chr6_+_44225233 | 0.00 |
ENSRNOT00000066593
|
Kidins220
|
kinase D-interacting substrate 220 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Aire
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071283 | response to high light intensity(GO:0009644) cellular response to iron(III) ion(GO:0071283) |
| 0.0 | 0.1 | GO:0009609 | response to symbiotic bacterium(GO:0009609) |
| 0.0 | 0.1 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.1 | GO:0098706 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.0 | GO:0052422 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0033590 | response to cobalamin(GO:0033590) |
| 0.0 | 0.0 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.0 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.0 | 0.0 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0090534 | calcium ion-transporting ATPase complex(GO:0090534) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.0 | GO:0032142 | dinucleotide insertion or deletion binding(GO:0032139) single guanine insertion binding(GO:0032142) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.0 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |