Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
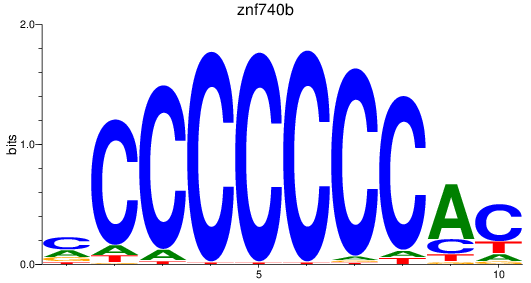
Results for znf740b
Z-value: 1.04
Transcription factors associated with znf740b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
znf740b
|
ENSDARG00000061174 | zinc finger protein 740b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| znf740b | dr11_v1_chr23_-_36418059_36418059 | 0.70 | 1.2e-03 | Click! |
Activity profile of znf740b motif
Sorted Z-values of znf740b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_1732838 | 4.39 |
ENSDART00000159555
ENSDART00000168161 |
FBLN1
|
fibulin 1 |
| chr2_+_22694382 | 4.10 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr3_-_16289826 | 3.52 |
ENSDART00000131972
|
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr2_-_44183613 | 3.13 |
ENSDART00000079596
|
cadm3
|
cell adhesion molecule 3 |
| chr17_-_4395373 | 3.07 |
ENSDART00000015923
|
klhl10a
|
kelch-like family member 10a |
| chr3_+_62126981 | 2.95 |
ENSDART00000060527
|
drc3
|
dynein regulatory complex subunit 3 |
| chr14_-_9522364 | 2.92 |
ENSDART00000054689
|
atoh8
|
atonal bHLH transcription factor 8 |
| chr2_-_44183451 | 2.89 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr10_-_5857548 | 2.83 |
ENSDART00000166933
|
si:ch211-281k19.2
|
si:ch211-281k19.2 |
| chr21_-_43117327 | 2.74 |
ENSDART00000122352
|
p4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide 2 |
| chr16_+_10963602 | 2.59 |
ENSDART00000141032
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr2_-_24907741 | 2.45 |
ENSDART00000155013
|
si:dkey-149i17.11
|
si:dkey-149i17.11 |
| chr18_-_41232297 | 2.37 |
ENSDART00000036928
|
fbxo36a
|
F-box protein 36a |
| chr23_-_524780 | 2.34 |
ENSDART00000055139
|
col9a3
|
collagen, type IX, alpha 3 |
| chr6_+_26314464 | 2.32 |
ENSDART00000115392
|
dzip1l
|
DAZ interacting zinc finger protein 1-like |
| chr15_-_5742531 | 2.29 |
ENSDART00000045985
|
phkg1a
|
phosphorylase kinase, gamma 1a (muscle) |
| chr5_-_68022631 | 2.22 |
ENSDART00000143199
|
wasf3a
|
WAS protein family, member 3a |
| chr16_+_10918252 | 2.20 |
ENSDART00000172949
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr10_+_6613279 | 2.16 |
ENSDART00000149643
|
si:ch211-57m13.1
|
si:ch211-57m13.1 |
| chr7_-_10606 | 2.01 |
ENSDART00000192650
ENSDART00000186761 |
FO704772.2
|
|
| chr25_+_150570 | 2.00 |
ENSDART00000170892
|
adam10b
|
ADAM metallopeptidase domain 10b |
| chr22_-_15957041 | 2.00 |
ENSDART00000149236
ENSDART00000187500 ENSDART00000176304 ENSDART00000080047 ENSDART00000190068 |
eps15l1a
|
epidermal growth factor receptor pathway substrate 15-like 1a |
| chr20_+_26538137 | 1.99 |
ENSDART00000045397
|
stx11b.1
|
syntaxin 11b, tandem duplicate 1 |
| chr8_-_23578660 | 1.97 |
ENSDART00000039080
|
wasb
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) b |
| chr20_-_7080427 | 1.93 |
ENSDART00000140166
ENSDART00000023677 |
si:ch211-121a2.2
|
si:ch211-121a2.2 |
| chr20_+_25340814 | 1.88 |
ENSDART00000063028
|
ctgfa
|
connective tissue growth factor a |
| chr19_-_6873107 | 1.87 |
ENSDART00000124440
|
CABZ01029822.1
|
|
| chr11_-_3334248 | 1.83 |
ENSDART00000154314
ENSDART00000121861 |
prph
|
peripherin |
| chr3_-_1190132 | 1.79 |
ENSDART00000149709
|
smdt1a
|
single-pass membrane protein with aspartate-rich tail 1a |
| chr25_-_18470695 | 1.78 |
ENSDART00000034377
|
cpa5
|
carboxypeptidase A5 |
| chr1_-_52498146 | 1.78 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr21_+_7823146 | 1.75 |
ENSDART00000030579
|
crhbp
|
corticotropin releasing hormone binding protein |
| chr10_+_8968203 | 1.74 |
ENSDART00000110443
ENSDART00000080772 |
fstb
|
follistatin b |
| chr22_+_35068046 | 1.74 |
ENSDART00000161660
ENSDART00000169573 |
si:ch73-173h19.3
|
si:ch73-173h19.3 |
| chr22_+_24157807 | 1.73 |
ENSDART00000159165
|
b3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr4_-_68563862 | 1.72 |
ENSDART00000182970
|
BX548011.2
|
|
| chr5_-_16351306 | 1.71 |
ENSDART00000168643
|
CABZ01088700.1
|
|
| chr24_+_42131564 | 1.65 |
ENSDART00000153854
|
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr2_-_26527730 | 1.62 |
ENSDART00000138693
|
si:ch211-106k21.5
|
si:ch211-106k21.5 |
| chr1_-_38709551 | 1.55 |
ENSDART00000128794
|
gpm6ab
|
glycoprotein M6Ab |
| chr24_-_28243186 | 1.54 |
ENSDART00000105691
|
cox4i1l
|
cytochrome c oxidase subunit IV isoform 1, like |
| chr10_+_10210455 | 1.54 |
ENSDART00000144214
|
sh2d3ca
|
SH2 domain containing 3Ca |
| chr13_-_45523026 | 1.53 |
ENSDART00000020663
|
rhd
|
Rh blood group, D antigen |
| chr18_+_21122818 | 1.52 |
ENSDART00000060015
ENSDART00000060184 |
chka
|
choline kinase alpha |
| chr7_-_35126374 | 1.51 |
ENSDART00000141211
|
hsd11b2
|
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr7_+_23495986 | 1.50 |
ENSDART00000190739
ENSDART00000115299 ENSDART00000101423 ENSDART00000142401 |
zgc:109889
|
zgc:109889 |
| chr13_-_8776474 | 1.45 |
ENSDART00000122371
|
STPG4
|
si:ch211-93n23.7 |
| chr2_-_23004286 | 1.44 |
ENSDART00000134664
ENSDART00000110373 ENSDART00000185833 ENSDART00000187235 |
znf414
mllt1b
|
zinc finger protein 414 MLLT1, super elongation complex subunit b |
| chr5_-_19052184 | 1.42 |
ENSDART00000133330
|
fam214b
|
family with sequence similarity 214, member B |
| chr3_-_59981162 | 1.42 |
ENSDART00000128790
|
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr24_+_42127983 | 1.42 |
ENSDART00000190157
ENSDART00000176032 ENSDART00000175790 |
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr24_-_21674950 | 1.42 |
ENSDART00000123216
ENSDART00000046211 |
lnx2a
|
ligand of numb-protein X 2a |
| chr15_-_5815006 | 1.42 |
ENSDART00000102459
|
rbp2a
|
retinol binding protein 2a, cellular |
| chr20_+_20672163 | 1.38 |
ENSDART00000027758
|
rtn1b
|
reticulon 1b |
| chr3_+_40809011 | 1.35 |
ENSDART00000033713
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr24_+_42132962 | 1.34 |
ENSDART00000187739
|
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr20_+_18740518 | 1.34 |
ENSDART00000142196
|
fam167ab
|
family with sequence similarity 167, member Ab |
| chr3_-_59981476 | 1.32 |
ENSDART00000035878
ENSDART00000124038 |
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr15_-_35246742 | 1.31 |
ENSDART00000131479
|
mff
|
mitochondrial fission factor |
| chr23_+_28494189 | 1.30 |
ENSDART00000146990
ENSDART00000006657 |
huwe1
|
HECT, UBA and WWE domain containing 1 |
| chr19_-_10043142 | 1.30 |
ENSDART00000193016
|
grin2da
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, a |
| chr2_+_24781026 | 1.29 |
ENSDART00000145692
|
pde4ca
|
phosphodiesterase 4C, cAMP-specific a |
| chr11_-_18705303 | 1.28 |
ENSDART00000059732
|
id1
|
inhibitor of DNA binding 1 |
| chr12_+_2522642 | 1.24 |
ENSDART00000152567
|
frmpd2
|
FERM and PDZ domain containing 2 |
| chr19_+_5315987 | 1.22 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr18_+_44649804 | 1.20 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr23_-_19715557 | 1.17 |
ENSDART00000143764
|
rpl10
|
ribosomal protein L10 |
| chr4_-_5597167 | 1.16 |
ENSDART00000132431
|
vegfab
|
vascular endothelial growth factor Ab |
| chr11_-_4298288 | 1.15 |
ENSDART00000188239
ENSDART00000183705 ENSDART00000187948 |
CT033790.1
|
|
| chr10_-_31563049 | 1.13 |
ENSDART00000023575
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr25_-_1124851 | 1.11 |
ENSDART00000067558
|
spg11
|
spastic paraplegia 11 |
| chr3_+_6469754 | 1.11 |
ENSDART00000185809
|
NUP85 (1 of many)
|
nucleoporin 85 |
| chr1_-_52497834 | 1.06 |
ENSDART00000136469
ENSDART00000004233 |
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr14_-_32089117 | 1.05 |
ENSDART00000158014
|
si:ch211-69b22.5
|
si:ch211-69b22.5 |
| chr1_-_52128425 | 1.04 |
ENSDART00000149939
|
rad23aa
|
RAD23 homolog A, nucleotide excision repair protein a |
| chr23_+_36083529 | 1.02 |
ENSDART00000053295
ENSDART00000130260 |
hoxc10a
|
homeobox C10a |
| chr2_+_33747509 | 1.02 |
ENSDART00000134187
|
si:dkey-31m5.5
|
si:dkey-31m5.5 |
| chr2_-_16565690 | 1.01 |
ENSDART00000022549
|
atp1b3a
|
ATPase Na+/K+ transporting subunit beta 3a |
| chr2_-_23411368 | 1.01 |
ENSDART00000159495
|
si:ch73-129a22.11
|
si:ch73-129a22.11 |
| chr10_+_2742499 | 1.00 |
ENSDART00000122847
|
grk5
|
G protein-coupled receptor kinase 5 |
| chr4_-_26108053 | 1.00 |
ENSDART00000066951
|
si:ch211-244b2.4
|
si:ch211-244b2.4 |
| chr5_-_3839285 | 0.99 |
ENSDART00000122292
|
mlxipl
|
MLX interacting protein like |
| chr5_+_6892195 | 0.97 |
ENSDART00000048201
|
gb:cr929477
|
expressed sequence CR929477 |
| chr15_+_3284684 | 0.97 |
ENSDART00000179778
|
foxo1a
|
forkhead box O1 a |
| chr9_+_2522797 | 0.97 |
ENSDART00000186786
ENSDART00000147034 |
gpr155a
|
G protein-coupled receptor 155a |
| chr16_-_1709328 | 0.97 |
ENSDART00000168865
|
ascc3
|
activating signal cointegrator 1 complex subunit 3 |
| chr8_+_8291492 | 0.94 |
ENSDART00000151314
|
srpk3
|
SRSF protein kinase 3 |
| chr1_+_144284 | 0.93 |
ENSDART00000064061
|
prozb
|
protein Z, vitamin K-dependent plasma glycoprotein b |
| chr16_+_9713850 | 0.92 |
ENSDART00000164103
|
ecm1b
|
extracellular matrix protein 1b |
| chr2_-_23443189 | 0.91 |
ENSDART00000168894
|
si:ch211-14p21.4
|
si:ch211-14p21.4 |
| chr11_-_497854 | 0.89 |
ENSDART00000104520
|
cnbpb
|
CCHC-type zinc finger, nucleic acid binding protein b |
| chr2_-_23479714 | 0.89 |
ENSDART00000167291
|
si:ch211-14p21.3
|
si:ch211-14p21.3 |
| chr12_-_22540943 | 0.89 |
ENSDART00000172310
|
zbtb4
|
zinc finger and BTB domain containing 4 |
| chr15_-_2734560 | 0.88 |
ENSDART00000153853
|
ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr12_-_7854216 | 0.87 |
ENSDART00000149594
|
ank3b
|
ankyrin 3b |
| chr19_+_22085925 | 0.87 |
ENSDART00000185636
|
atp9b
|
ATPase phospholipid transporting 9B |
| chr17_+_3379673 | 0.87 |
ENSDART00000176354
|
sntg2
|
syntrophin, gamma 2 |
| chr2_+_23790748 | 0.86 |
ENSDART00000041877
|
csrnp1a
|
cysteine-serine-rich nuclear protein 1a |
| chr13_-_31435137 | 0.86 |
ENSDART00000057441
|
rtn1a
|
reticulon 1a |
| chr15_+_3284416 | 0.85 |
ENSDART00000187665
ENSDART00000171723 |
foxo1a
|
forkhead box O1 a |
| chr24_+_41931585 | 0.85 |
ENSDART00000130310
|
epb41l3a
|
erythrocyte membrane protein band 4.1-like 3a |
| chr7_+_1449999 | 0.85 |
ENSDART00000173864
|
si:cabz01101003.1
|
si:cabz01101003.1 |
| chr7_-_48665305 | 0.84 |
ENSDART00000190507
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr3_-_33417826 | 0.84 |
ENSDART00000084284
|
abi3a
|
ABI family, member 3a |
| chr4_-_26107841 | 0.83 |
ENSDART00000172012
|
si:ch211-244b2.4
|
si:ch211-244b2.4 |
| chr25_-_11088839 | 0.82 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr15_-_30832897 | 0.82 |
ENSDART00000152330
|
msi2b
|
musashi RNA-binding protein 2b |
| chr12_-_47899497 | 0.80 |
ENSDART00000162219
|
QRFPR
|
pyroglutamylated RFamide peptide receptor |
| chr20_-_5052786 | 0.79 |
ENSDART00000138818
ENSDART00000181655 ENSDART00000164274 |
arid1b
|
AT rich interactive domain 1B (SWI1-like) |
| chr2_+_3595333 | 0.79 |
ENSDART00000041052
|
c1ql3b
|
complement component 1, q subcomponent-like 3b |
| chr23_-_35064785 | 0.78 |
ENSDART00000172240
|
BX294434.1
|
|
| chr2_-_23515584 | 0.78 |
ENSDART00000170748
|
si:dkey-58b18.5
|
si:dkey-58b18.5 |
| chr2_-_23577672 | 0.78 |
ENSDART00000132003
|
si:dkey-58b18.5
|
si:dkey-58b18.5 |
| chr21_-_308852 | 0.76 |
ENSDART00000151613
|
lhfpl2a
|
LHFPL tetraspan subfamily member 2a |
| chr5_+_30699260 | 0.76 |
ENSDART00000012888
|
atp5l
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit g |
| chr9_+_13714379 | 0.75 |
ENSDART00000017593
ENSDART00000145503 |
tmem237a
|
transmembrane protein 237a |
| chr2_-_17694504 | 0.75 |
ENSDART00000133709
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr2_+_33052360 | 0.73 |
ENSDART00000134651
|
rnf220a
|
ring finger protein 220a |
| chr16_-_41439659 | 0.73 |
ENSDART00000191624
|
cpne4a
|
copine IVa |
| chr2_-_6051836 | 0.73 |
ENSDART00000092479
|
si:ch211-284b7.3
|
si:ch211-284b7.3 |
| chr2_-_23546627 | 0.73 |
ENSDART00000141342
|
si:dkey-58b18.2
|
si:dkey-58b18.2 |
| chr8_+_46010838 | 0.73 |
ENSDART00000143126
|
si:ch211-119d14.2
|
si:ch211-119d14.2 |
| chr23_+_36101185 | 0.72 |
ENSDART00000103139
|
hoxc8a
|
homeobox C8a |
| chr20_-_51727860 | 0.72 |
ENSDART00000147044
|
brox
|
BRO1 domain and CAAX motif containing |
| chr7_+_46261199 | 0.72 |
ENSDART00000170390
ENSDART00000183227 |
znf536
|
zinc finger protein 536 |
| chr17_+_53418445 | 0.72 |
ENSDART00000097631
|
slc9a1b
|
solute carrier family 9 member A1b |
| chr1_-_46632948 | 0.69 |
ENSDART00000148893
ENSDART00000053232 |
cdadc1
|
cytidine and dCMP deaminase domain containing 1 |
| chr11_-_7078392 | 0.68 |
ENSDART00000112156
ENSDART00000188556 |
si:ch211-253b8.5
|
si:ch211-253b8.5 |
| chr16_-_23800484 | 0.67 |
ENSDART00000139964
|
rps27.2
|
ribosomal protein S27, isoform 2 |
| chr20_-_20610812 | 0.66 |
ENSDART00000181870
|
ppm1ab
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Ab |
| chr21_+_25660613 | 0.65 |
ENSDART00000134017
|
si:dkey-17e16.15
|
si:dkey-17e16.15 |
| chr5_-_40510397 | 0.63 |
ENSDART00000146237
ENSDART00000051065 |
fsta
|
follistatin a |
| chr2_+_33052169 | 0.63 |
ENSDART00000180008
|
rnf220a
|
ring finger protein 220a |
| chr2_+_7192966 | 0.63 |
ENSDART00000142735
|
si:ch211-13f8.1
|
si:ch211-13f8.1 |
| chr8_+_23711842 | 0.62 |
ENSDART00000128783
|
ppardb
|
peroxisome proliferator-activated receptor delta b |
| chr25_-_37501371 | 0.62 |
ENSDART00000160498
|
CABZ01088346.1
|
|
| chr23_+_5977965 | 0.61 |
ENSDART00000115403
ENSDART00000183147 |
NAV1 (1 of many)
|
neuron navigator 1 |
| chr1_+_39553040 | 0.60 |
ENSDART00000137676
|
tenm3
|
teneurin transmembrane protein 3 |
| chr2_+_36004381 | 0.59 |
ENSDART00000098706
|
lamc2
|
laminin, gamma 2 |
| chr18_-_50845804 | 0.58 |
ENSDART00000158517
|
PDPR
|
si:cabz01113374.3 |
| chr17_-_8976307 | 0.57 |
ENSDART00000092113
|
zranb1b
|
zinc finger, RAN-binding domain containing 1b |
| chr18_+_14529005 | 0.57 |
ENSDART00000186379
|
kcng4a
|
potassium voltage-gated channel, subfamily G, member 4a |
| chr18_-_7810214 | 0.56 |
ENSDART00000139505
ENSDART00000139188 |
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr9_-_31278048 | 0.56 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr25_+_35502552 | 0.55 |
ENSDART00000189612
ENSDART00000058443 |
fibina
|
fin bud initiation factor a |
| chr7_-_55454406 | 0.54 |
ENSDART00000108646
|
piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr23_+_32499916 | 0.53 |
ENSDART00000134811
|
si:dkey-261h17.1
|
si:dkey-261h17.1 |
| chr3_+_23703704 | 0.52 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr24_-_28381404 | 0.52 |
ENSDART00000148406
|
prkag2a
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit a |
| chr18_+_17827149 | 0.51 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr18_+_35742838 | 0.51 |
ENSDART00000088504
ENSDART00000140386 |
rasgrp4
|
RAS guanyl releasing protein 4 |
| chr2_+_16487443 | 0.51 |
ENSDART00000114980
|
CR377211.1
|
|
| chr3_+_37824268 | 0.50 |
ENSDART00000137038
|
asic2
|
acid-sensing (proton-gated) ion channel 2 |
| chr6_+_15373153 | 0.50 |
ENSDART00000155865
|
tmtops2a
|
teleost multiple tissue opsin 2a |
| chr8_+_25267903 | 0.50 |
ENSDART00000093090
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr1_-_51474974 | 0.49 |
ENSDART00000152719
|
spred2a
|
sprouty-related, EVH1 domain containing 2a |
| chr3_+_52467879 | 0.49 |
ENSDART00000156039
|
adgre5a
|
adhesion G protein-coupled receptor E5a |
| chr15_+_2534740 | 0.48 |
ENSDART00000138469
|
cux1b
|
cut-like homeobox 1b |
| chr11_+_6650966 | 0.48 |
ENSDART00000131236
|
si:dkey-246j7.1
|
si:dkey-246j7.1 |
| chr3_-_26204867 | 0.47 |
ENSDART00000103748
|
gdpd3a
|
glycerophosphodiester phosphodiesterase domain containing 3a |
| chr4_+_34417403 | 0.47 |
ENSDART00000186032
ENSDART00000167909 |
si:ch211-246b8.2
|
si:ch211-246b8.2 |
| chr7_+_41314862 | 0.46 |
ENSDART00000185198
|
zgc:165532
|
zgc:165532 |
| chr7_+_8361083 | 0.46 |
ENSDART00000102091
|
jac7
|
jacalin 7 |
| chr2_+_54389750 | 0.46 |
ENSDART00000189236
|
rab12
|
RAB12, member RAS oncogene family |
| chr16_-_31435020 | 0.46 |
ENSDART00000138508
|
zgc:194210
|
zgc:194210 |
| chr11_-_41130239 | 0.45 |
ENSDART00000173268
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr2_-_16217344 | 0.44 |
ENSDART00000152031
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr7_-_69636502 | 0.44 |
ENSDART00000126739
|
tspan5a
|
tetraspanin 5a |
| chr13_+_771403 | 0.44 |
ENSDART00000093166
|
nrxn1b
|
neurexin 1b |
| chr25_+_4750972 | 0.43 |
ENSDART00000168903
|
si:zfos-2372e4.1
|
si:zfos-2372e4.1 |
| chr3_+_13440900 | 0.43 |
ENSDART00000143715
|
GNG14
|
si:dkey-117i10.1 |
| chr21_+_30351256 | 0.41 |
ENSDART00000078341
|
foxi3a
|
forkhead box I3a |
| chr22_+_17261801 | 0.41 |
ENSDART00000192978
ENSDART00000193187 ENSDART00000179953 ENSDART00000134798 |
tdrd5
|
tudor domain containing 5 |
| chr18_+_45990394 | 0.41 |
ENSDART00000024068
|
mmp23bb
|
matrix metallopeptidase 23bb |
| chr25_+_13191615 | 0.40 |
ENSDART00000168849
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr6_-_43677125 | 0.40 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr21_-_11820379 | 0.39 |
ENSDART00000126640
|
RHOBTB3
|
si:dkey-6b12.5 |
| chr9_-_39968820 | 0.39 |
ENSDART00000100311
|
IKZF2
|
si:zfos-1425h8.1 |
| chr9_+_29585943 | 0.38 |
ENSDART00000185989
ENSDART00000115290 |
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr23_-_9864166 | 0.38 |
ENSDART00000124510
ENSDART00000187934 |
prkcbp1l
|
protein kinase C binding protein 1, like |
| chr7_+_36552725 | 0.38 |
ENSDART00000173682
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr20_-_25542183 | 0.38 |
ENSDART00000024350
|
cyp2ad2
|
cytochrome P450, family 2, subfamily AD, polypeptide 2 |
| chr19_-_46957968 | 0.38 |
ENSDART00000043713
|
angpt1
|
angiopoietin 1 |
| chr25_+_13191391 | 0.38 |
ENSDART00000109937
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr24_-_40860603 | 0.38 |
ENSDART00000188032
|
CU633479.7
|
|
| chr15_-_40267485 | 0.37 |
ENSDART00000152253
|
kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr1_-_44638058 | 0.37 |
ENSDART00000081835
|
slc43a1b
|
solute carrier family 43 (amino acid system L transporter), member 1b |
| chr19_+_42983613 | 0.37 |
ENSDART00000033724
|
fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr1_+_31112436 | 0.36 |
ENSDART00000075340
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr23_+_42434348 | 0.36 |
ENSDART00000161027
|
cyp2aa1
|
cytochrome P450, family 2, subfamily AA, polypeptide 1 |
| chr13_+_23677949 | 0.36 |
ENSDART00000144215
|
pcnxl2
|
pecanex-like 2 (Drosophila) |
| chr3_-_31804481 | 0.36 |
ENSDART00000028270
|
gfap
|
glial fibrillary acidic protein |
| chr7_+_38249858 | 0.35 |
ENSDART00000150158
|
si:dkey-78a14.4
|
si:dkey-78a14.4 |
| chr18_-_16181952 | 0.35 |
ENSDART00000157824
|
slc6a15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr10_-_43113731 | 0.35 |
ENSDART00000138099
|
tmem167a
|
transmembrane protein 167A |
| chr4_-_73572030 | 0.34 |
ENSDART00000121652
|
znf1015
|
zinc finger protein 1015 |
| chr23_+_44049509 | 0.34 |
ENSDART00000102003
|
txk
|
TXK tyrosine kinase |
| chr17_+_52822422 | 0.33 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr7_+_61480296 | 0.33 |
ENSDART00000083255
|
adam19a
|
ADAM metallopeptidase domain 19a |
| chr11_-_40504170 | 0.32 |
ENSDART00000165394
|
si:dkeyp-61b2.1
|
si:dkeyp-61b2.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of znf740b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.6 | 1.8 | GO:0002792 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.4 | 1.2 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.4 | 5.7 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.3 | 0.9 | GO:0060031 | mediolateral intercalation(GO:0060031) |
| 0.3 | 1.8 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.2 | 2.4 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 1.5 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.2 | 1.5 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.2 | 0.9 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.2 | 0.6 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.2 | 1.4 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.1 | 0.6 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 2.7 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 2.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 1.7 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.8 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 1.0 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 1.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.2 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 0.6 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.5 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.1 | 1.3 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 0.4 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 0.4 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.1 | 0.3 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.8 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 0.1 | 0.8 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.5 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 1.1 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 0.6 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 0.3 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 0.7 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.5 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 0.6 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.1 | 2.0 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 0.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 1.3 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 5.9 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.1 | 2.0 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 1.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 1.0 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 0.3 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.1 | 0.8 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.1 | 1.1 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 2.3 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.4 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 0.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.9 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 1.1 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.8 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.7 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 1.3 | GO:0045103 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.3 | GO:0042664 | locus ceruleus development(GO:0021703) negative regulation of endodermal cell fate specification(GO:0042664) |
| 0.0 | 0.2 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 1.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.5 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.2 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 1.0 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.4 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 6.5 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.2 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 2.8 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 0.1 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.0 | 0.2 | GO:0030816 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.5 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.2 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.1 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.2 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.2 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.6 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 0.3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.3 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.0 | 0.6 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.9 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 2.0 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 0.1 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.3 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.6 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 2.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 1.8 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 1.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 1.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 2.0 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.1 | 1.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 2.0 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 2.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0043514 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.0 | 0.9 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 2.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 3.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.3 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 3.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.8 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 1.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.7 | GO:0022627 | cytosolic ribosome(GO:0022626) cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.5 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.8 | GO:0030027 | lamellipodium(GO:0030027) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.6 | 1.7 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.5 | 2.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.4 | 3.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.3 | 1.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.3 | 2.8 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.2 | 2.7 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.2 | 3.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 1.7 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 2.4 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 3.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.7 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.6 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.3 | GO:0072571 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.1 | 1.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.3 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 1.0 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.3 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 1.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.7 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.4 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 1.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.2 | GO:2001070 | starch binding(GO:2001070) |
| 0.1 | 0.3 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 1.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.8 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 1.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.5 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 1.0 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0042164 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.0 | 3.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.1 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 2.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 8.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.5 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 2.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.9 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 1.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 1.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 4.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.3 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 2.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 4.7 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.0 | 0.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.6 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 1.0 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.6 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |