Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
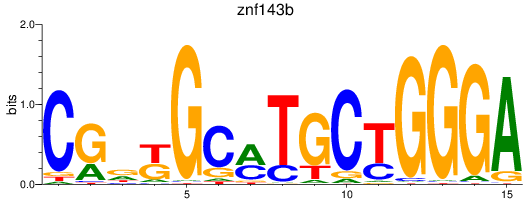
Results for znf143b
Z-value: 0.34
Transcription factors associated with znf143b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
znf143b
|
ENSDARG00000041581 | zinc finger protein 143b |
|
znf143b
|
ENSDARG00000111018 | zinc finger protein 143b |
|
znf143b
|
ENSDARG00000112408 | zinc finger protein 143b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| znf143b | dr11_v1_chr18_-_16937008_16937109 | 0.91 | 1.3e-07 | Click! |
Activity profile of znf143b motif
Sorted Z-values of znf143b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_43636595 | 1.05 |
ENSDART00000151115
ENSDART00000151486 ENSDART00000151778 |
si:ch1073-263o8.2
|
si:ch1073-263o8.2 |
| chr10_-_8079737 | 0.82 |
ENSDART00000059014
ENSDART00000179549 |
zgc:173443
si:ch211-251f6.6
|
zgc:173443 si:ch211-251f6.6 |
| chr3_-_3448095 | 0.70 |
ENSDART00000078886
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr9_-_1639884 | 0.56 |
ENSDART00000062850
ENSDART00000150947 |
agps
|
alkylglycerone phosphate synthase |
| chr17_+_46387086 | 0.53 |
ENSDART00000157079
|
si:dkey-206p8.1
|
si:dkey-206p8.1 |
| chr23_+_7692042 | 0.51 |
ENSDART00000018512
|
pofut1
|
protein O-fucosyltransferase 1 |
| chr16_+_31511739 | 0.50 |
ENSDART00000049420
|
ndrg1b
|
N-myc downstream regulated 1b |
| chr17_+_22760126 | 0.50 |
ENSDART00000151999
ENSDART00000190442 |
ttc27
|
tetratricopeptide repeat domain 27 |
| chr15_-_47193564 | 0.45 |
ENSDART00000172453
|
LSAMP
|
limbic system-associated membrane protein |
| chr12_-_1034383 | 0.45 |
ENSDART00000152455
ENSDART00000152346 |
polr3e
|
polymerase (RNA) III (DNA directed) polypeptide E |
| chr9_+_14028157 | 0.43 |
ENSDART00000134603
|
si:ch211-67e16.4
|
si:ch211-67e16.4 |
| chr9_-_8297344 | 0.42 |
ENSDART00000180945
|
si:ch211-145c1.1
|
si:ch211-145c1.1 |
| chr9_-_55946377 | 0.37 |
ENSDART00000161536
ENSDART00000169432 |
SH3RF3
|
si:ch211-124n19.2 |
| chr8_+_54263946 | 0.37 |
ENSDART00000193119
|
TMCC1 (1 of many)
|
transmembrane and coiled-coil domain family 1 |
| chr21_+_7131970 | 0.36 |
ENSDART00000161921
|
zgc:113019
|
zgc:113019 |
| chr21_+_7605803 | 0.35 |
ENSDART00000121813
|
wdr41
|
WD repeat domain 41 |
| chr25_+_8925934 | 0.35 |
ENSDART00000073914
|
accs
|
1-aminocyclopropane-1-carboxylate synthase homolog (Arabidopsis)(non-functional) |
| chr9_-_11676491 | 0.35 |
ENSDART00000022358
|
zc3h15
|
zinc finger CCCH-type containing 15 |
| chr15_-_41245962 | 0.34 |
ENSDART00000155359
|
smco4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr15_-_28805493 | 0.34 |
ENSDART00000179617
|
cd3eap
|
CD3e molecule, epsilon associated protein |
| chr4_-_9780931 | 0.34 |
ENSDART00000134280
ENSDART00000150664 ENSDART00000150304 ENSDART00000080744 |
svopl
|
SVOP-like |
| chr19_-_34970312 | 0.34 |
ENSDART00000102896
|
ndrg1a
|
N-myc downstream regulated 1a |
| chr18_+_18000887 | 0.33 |
ENSDART00000147797
|
si:ch211-212o1.2
|
si:ch211-212o1.2 |
| chr3_+_1219344 | 0.33 |
ENSDART00000161945
|
rrp7a
|
ribosomal RNA processing 7 homolog A |
| chr19_+_40069524 | 0.33 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chr3_-_34816893 | 0.32 |
ENSDART00000084448
ENSDART00000154696 |
psmd11a
|
proteasome 26S subunit, non-ATPase 11a |
| chr13_-_51846224 | 0.32 |
ENSDART00000184663
|
LT631684.2
|
|
| chr24_-_26622423 | 0.31 |
ENSDART00000182044
|
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr7_-_8344243 | 0.31 |
ENSDART00000149510
|
f13a1b
|
coagulation factor XIII, A1 polypeptide b |
| chr3_+_35608385 | 0.31 |
ENSDART00000193219
ENSDART00000132703 |
traf7
|
TNF receptor-associated factor 7 |
| chr18_-_8380090 | 0.30 |
ENSDART00000141581
ENSDART00000081143 |
sephs1
|
selenophosphate synthetase 1 |
| chr4_+_14981854 | 0.30 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr15_-_1622468 | 0.28 |
ENSDART00000149008
ENSDART00000034456 |
kpna4
|
karyopherin alpha 4 (importin alpha 3) |
| chr15_+_24676905 | 0.27 |
ENSDART00000078014
ENSDART00000143137 |
poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr19_-_7358184 | 0.26 |
ENSDART00000092379
|
oxr1b
|
oxidation resistance 1b |
| chr14_+_32852388 | 0.26 |
ENSDART00000166351
|
nkrf
|
NFKB repressing factor |
| chr14_-_30876708 | 0.25 |
ENSDART00000147597
|
ubl3b
|
ubiquitin-like 3b |
| chr14_+_16287968 | 0.24 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr2_-_1227221 | 0.24 |
ENSDART00000130897
|
ABCF3
|
ATP binding cassette subfamily F member 3 |
| chr18_+_18000695 | 0.24 |
ENSDART00000146898
|
si:ch211-212o1.2
|
si:ch211-212o1.2 |
| chr5_+_30089715 | 0.24 |
ENSDART00000078114
|
timm8b
|
translocase of inner mitochondrial membrane 8 homolog B (yeast) |
| chr16_-_8132742 | 0.23 |
ENSDART00000104323
|
snrka
|
SNF related kinase a |
| chr3_-_50066499 | 0.23 |
ENSDART00000056618
ENSDART00000154561 |
mrpl12
|
mitochondrial ribosomal protein L12 |
| chr3_-_40301467 | 0.23 |
ENSDART00000055186
|
atp5mf
|
ATP synthase membrane subunit f |
| chr2_-_34138064 | 0.23 |
ENSDART00000133381
|
cenpl
|
centromere protein L |
| chr1_+_53279861 | 0.22 |
ENSDART00000035713
|
rnf150a
|
ring finger protein 150a |
| chr15_-_6966221 | 0.22 |
ENSDART00000165487
ENSDART00000027657 |
mrps22
|
mitochondrial ribosomal protein S22 |
| chr9_+_26086135 | 0.22 |
ENSDART00000011910
|
arglu1a
|
arginine and glutamate rich 1a |
| chr4_-_4834617 | 0.22 |
ENSDART00000141539
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr18_+_49969568 | 0.22 |
ENSDART00000126916
|
mob2b
|
MOB kinase activator 2b |
| chr13_-_46687097 | 0.21 |
ENSDART00000169106
ENSDART00000158202 |
CABZ01078449.1
|
|
| chr5_-_32383475 | 0.21 |
ENSDART00000141294
|
fbxw2
|
F-box and WD repeat domain containing 2 |
| chr4_-_4834347 | 0.21 |
ENSDART00000141803
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr19_+_43715911 | 0.21 |
ENSDART00000006344
|
cap1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr7_-_62003831 | 0.21 |
ENSDART00000113585
ENSDART00000062704 |
plaa
|
phospholipase A2-activating protein |
| chr14_-_30876299 | 0.21 |
ENSDART00000180305
|
ubl3b
|
ubiquitin-like 3b |
| chr10_-_40826657 | 0.20 |
ENSDART00000076304
|
pcna
|
proliferating cell nuclear antigen |
| chr19_-_33996277 | 0.20 |
ENSDART00000159384
|
elmo1
|
engulfment and cell motility 1 (ced-12 homolog, C. elegans) |
| chr16_+_19637384 | 0.20 |
ENSDART00000184773
ENSDART00000191895 ENSDART00000182020 ENSDART00000135359 |
macc1
|
metastasis associated in colon cancer 1 |
| chr2_-_37537887 | 0.20 |
ENSDART00000143496
ENSDART00000025841 |
arhgef18a
|
rho/rac guanine nucleotide exchange factor (GEF) 18a |
| chr14_+_32926385 | 0.19 |
ENSDART00000139159
|
lnx2b
|
ligand of numb-protein X 2b |
| chr10_-_35186310 | 0.19 |
ENSDART00000127805
|
pom121
|
POM121 transmembrane nucleoporin |
| chr19_+_15521997 | 0.19 |
ENSDART00000003164
|
ppp1r8a
|
protein phosphatase 1, regulatory subunit 8a |
| chr17_+_37253706 | 0.19 |
ENSDART00000076004
|
tmem62
|
transmembrane protein 62 |
| chr4_+_288633 | 0.19 |
ENSDART00000183304
|
FO834823.1
|
|
| chr5_+_34549845 | 0.19 |
ENSDART00000139317
|
aif1l
|
allograft inflammatory factor 1-like |
| chr15_+_44184367 | 0.19 |
ENSDART00000162918
ENSDART00000110060 |
zgc:165514
|
zgc:165514 |
| chr3_+_18050667 | 0.19 |
ENSDART00000035531
|
mettl26
|
methyltransferase like 26 |
| chr13_+_30506781 | 0.19 |
ENSDART00000110884
|
zmiz1a
|
zinc finger, MIZ-type containing 1a |
| chr21_+_30007566 | 0.19 |
ENSDART00000149124
ENSDART00000150071 |
ttc1
|
tetratricopeptide repeat domain 1 |
| chr1_-_21563040 | 0.19 |
ENSDART00000049572
|
ncapd3
|
non-SMC condensin II complex, subunit D3 |
| chr16_+_30604387 | 0.19 |
ENSDART00000058785
|
fam210ab
|
family with sequence similarity 210, member Ab |
| chr15_-_9257136 | 0.19 |
ENSDART00000187901
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr6_+_12482599 | 0.19 |
ENSDART00000090316
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr22_+_20172018 | 0.18 |
ENSDART00000188104
|
hmg20b
|
high mobility group 20B |
| chr6_-_6993046 | 0.18 |
ENSDART00000053304
|
si:ch211-114n24.6
|
si:ch211-114n24.6 |
| chr2_+_31437547 | 0.17 |
ENSDART00000141170
|
stam
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr22_+_38919265 | 0.17 |
ENSDART00000148051
|
senp5
|
SUMO1/sentrin specific peptidase 5 |
| chr16_+_46401756 | 0.17 |
ENSDART00000147370
ENSDART00000144000 |
rpz2
|
rapunzel 2 |
| chr10_-_27009413 | 0.17 |
ENSDART00000139942
ENSDART00000146983 ENSDART00000132352 |
uqcc3
|
ubiquinol-cytochrome c reductase complex assembly factor 3 |
| chr23_-_4019699 | 0.17 |
ENSDART00000159780
|
slc9a8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr17_-_14780578 | 0.16 |
ENSDART00000154690
|
si:ch211-266o15.1
|
si:ch211-266o15.1 |
| chr23_-_4019928 | 0.16 |
ENSDART00000021062
|
slc9a8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr22_-_506522 | 0.15 |
ENSDART00000106645
ENSDART00000067637 |
dstyk
|
dual serine/threonine and tyrosine protein kinase |
| chr19_+_31532043 | 0.15 |
ENSDART00000136289
|
tmem64
|
transmembrane protein 64 |
| chr9_+_20869166 | 0.15 |
ENSDART00000147892
|
wdr3
|
WD repeat domain 3 |
| chr4_+_1619584 | 0.15 |
ENSDART00000148486
|
scaf11
|
SR-related CTD-associated factor 11 |
| chr16_-_39267185 | 0.15 |
ENSDART00000058550
ENSDART00000133642 |
gpd1l
|
glycerol-3-phosphate dehydrogenase 1 like |
| chr15_+_25452092 | 0.14 |
ENSDART00000009545
|
pak4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr19_-_12648122 | 0.14 |
ENSDART00000151184
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr5_-_68641353 | 0.14 |
ENSDART00000048432
|
dlg4a
|
discs, large homolog 4a (Drosophila) |
| chr5_+_23136544 | 0.13 |
ENSDART00000003428
ENSDART00000109340 ENSDART00000171039 ENSDART00000178821 |
prps1a
|
phosphoribosyl pyrophosphate synthetase 1A |
| chr13_-_27620815 | 0.13 |
ENSDART00000139904
|
kcnq5a
|
potassium voltage-gated channel, KQT-like subfamily, member 5a |
| chr10_-_36618674 | 0.13 |
ENSDART00000135302
|
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 |
| chr25_+_12012 | 0.13 |
ENSDART00000170348
|
cntn1a
|
contactin 1a |
| chr7_-_34184282 | 0.13 |
ENSDART00000183284
|
smad6a
|
SMAD family member 6a |
| chr6_-_17999776 | 0.13 |
ENSDART00000183048
ENSDART00000181577 ENSDART00000170597 |
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr2_-_34138400 | 0.13 |
ENSDART00000056667
|
cenpl
|
centromere protein L |
| chr5_+_34549365 | 0.12 |
ENSDART00000009500
|
aif1l
|
allograft inflammatory factor 1-like |
| chr3_-_18384501 | 0.12 |
ENSDART00000027630
|
kdelr2a
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2a |
| chr2_+_47718605 | 0.12 |
ENSDART00000189180
ENSDART00000148824 |
mbnl1
|
muscleblind-like splicing regulator 1 |
| chr20_-_9428021 | 0.12 |
ENSDART00000025330
|
rdh14b
|
retinol dehydrogenase 14b |
| chr10_-_36633882 | 0.12 |
ENSDART00000077161
ENSDART00000137688 |
rsf1b.1
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 remodeling and spacing factor 1b, tandem duplicate 1 |
| chr25_-_13871118 | 0.12 |
ENSDART00000160866
|
cry2
|
cryptochrome circadian clock 2 |
| chr5_-_13353810 | 0.11 |
ENSDART00000146136
ENSDART00000099602 ENSDART00000004539 |
ube2l3b
|
ubiquitin-conjugating enzyme E2L 3b |
| chr23_+_33947874 | 0.11 |
ENSDART00000136104
|
si:ch211-148l7.4
|
si:ch211-148l7.4 |
| chr8_+_24299321 | 0.11 |
ENSDART00000112046
ENSDART00000183558 ENSDART00000183796 ENSDART00000181395 ENSDART00000184555 ENSDART00000172531 ENSDART00000191663 ENSDART00000184103 ENSDART00000183676 |
ZNF335
|
si:ch211-269m15.3 |
| chr20_-_15161669 | 0.11 |
ENSDART00000080333
ENSDART00000063882 |
plpp6
|
phospholipid phosphatase 6 |
| chr8_+_21254192 | 0.11 |
ENSDART00000167718
|
itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr22_-_24248420 | 0.11 |
ENSDART00000165433
|
rgs2
|
regulator of G protein signaling 2 |
| chr17_-_6382392 | 0.11 |
ENSDART00000188051
ENSDART00000192560 ENSDART00000137389 ENSDART00000115389 |
txlnbb
|
taxilin beta b |
| chr7_+_25103965 | 0.11 |
ENSDART00000192561
ENSDART00000173721 |
si:dkey-23i12.9
|
si:dkey-23i12.9 |
| chr3_-_54846444 | 0.11 |
ENSDART00000074010
|
ubald1b
|
UBA-like domain containing 1b |
| chr25_-_37227491 | 0.10 |
ENSDART00000156647
|
glg1a
|
golgi glycoprotein 1a |
| chr2_-_30912307 | 0.10 |
ENSDART00000188653
|
myl12.2
|
myosin, light chain 12, genome duplicate 2 |
| chr16_+_42471455 | 0.10 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr13_-_49819027 | 0.10 |
ENSDART00000067824
|
b3galnt2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr2_-_23349116 | 0.10 |
ENSDART00000099690
|
fam129ab
|
family with sequence similarity 129, member Ab |
| chr22_+_26600834 | 0.10 |
ENSDART00000157411
|
adcy9
|
adenylate cyclase 9 |
| chr16_-_34212912 | 0.10 |
ENSDART00000145017
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr11_-_24532988 | 0.10 |
ENSDART00000067078
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr7_+_50766094 | 0.10 |
ENSDART00000165037
|
si:ch73-380l10.2
|
si:ch73-380l10.2 |
| chr9_+_27354653 | 0.10 |
ENSDART00000134134
|
tlr20.2
|
toll-like receptor 20, tandem duplicate 2 |
| chr24_+_39129316 | 0.09 |
ENSDART00000155346
|
tbc1d24
|
TBC1 domain family, member 24 |
| chr23_+_44263986 | 0.09 |
ENSDART00000149194
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr1_-_49950643 | 0.09 |
ENSDART00000138301
|
sgms2
|
sphingomyelin synthase 2 |
| chr16_-_54187374 | 0.09 |
ENSDART00000186989
ENSDART00000190113 |
LO017872.1
|
|
| chr5_+_20148671 | 0.09 |
ENSDART00000143205
|
svopa
|
SV2 related protein a |
| chr22_-_18779232 | 0.09 |
ENSDART00000186726
|
atp5f1d
|
ATP synthase F1 subunit delta |
| chr21_+_4320769 | 0.08 |
ENSDART00000168553
|
HTRA2 (1 of many)
|
si:dkey-84o3.4 |
| chr17_+_23470967 | 0.08 |
ENSDART00000104718
ENSDART00000154716 |
kif20ba
|
kinesin family member 20Ba |
| chr25_-_25646806 | 0.08 |
ENSDART00000089066
|
tbc1d2b
|
TBC1 domain family, member 2B |
| chr22_-_10774735 | 0.08 |
ENSDART00000081156
|
timm13
|
translocase of inner mitochondrial membrane 13 homolog (yeast) |
| chr2_-_12242695 | 0.08 |
ENSDART00000158175
|
gpr158b
|
G protein-coupled receptor 158b |
| chr11_+_44617021 | 0.08 |
ENSDART00000158887
|
rbm34
|
RNA binding motif protein 34 |
| chr19_-_6631900 | 0.08 |
ENSDART00000144571
|
pvrl2l
|
poliovirus receptor-related 2 like |
| chr20_-_15161502 | 0.08 |
ENSDART00000187072
|
plpp6
|
phospholipid phosphatase 6 |
| chr2_-_13691834 | 0.08 |
ENSDART00000186570
|
CABZ01044235.1
|
|
| chr5_-_23583446 | 0.08 |
ENSDART00000027217
|
cwc15
|
CWC15 spliceosome-associated protein homolog (S. cerevisiae) |
| chr3_+_29179329 | 0.08 |
ENSDART00000085216
ENSDART00000190136 |
cacna1i
|
calcium channel, voltage-dependent, T type, alpha 1I subunit |
| chr9_+_27379193 | 0.08 |
ENSDART00000142656
|
tlr20.4
|
toll-like receptor 20, tandem duplicate 4 |
| chr22_-_18778988 | 0.08 |
ENSDART00000019235
|
atp5f1d
|
ATP synthase F1 subunit delta |
| chr10_+_37182626 | 0.08 |
ENSDART00000137636
|
ksr1a
|
kinase suppressor of ras 1a |
| chr24_-_31092711 | 0.07 |
ENSDART00000168398
|
hccsb
|
holocytochrome c synthase b |
| chr9_+_33158191 | 0.07 |
ENSDART00000180786
|
dopey2
|
dopey family member 2 |
| chr14_+_41409697 | 0.07 |
ENSDART00000173335
|
bcorl1
|
BCL6 corepressor-like 1 |
| chr11_+_19060079 | 0.07 |
ENSDART00000158123
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr14_+_989733 | 0.07 |
ENSDART00000161487
ENSDART00000127317 |
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr17_+_28706946 | 0.07 |
ENSDART00000126967
|
strn3
|
striatin, calmodulin binding protein 3 |
| chr17_-_14671098 | 0.07 |
ENSDART00000037371
|
ppp1r13ba
|
protein phosphatase 1, regulatory subunit 13Ba |
| chr5_-_15321451 | 0.06 |
ENSDART00000139203
|
txnrd2.1
|
thioredoxin reductase 2, tandem duplicate 1 |
| chr2_+_53357953 | 0.06 |
ENSDART00000187577
|
celf5b
|
cugbp, Elav-like family member 5b |
| chr22_-_5918670 | 0.06 |
ENSDART00000141373
|
si:rp71-36a1.1
|
si:rp71-36a1.1 |
| chr22_-_2929127 | 0.06 |
ENSDART00000123730
ENSDART00000046678 |
pak2b
|
p21 protein (Cdc42/Rac)-activated kinase 2b |
| chr20_+_44498056 | 0.06 |
ENSDART00000023763
|
wdcp
|
WD repeat and coiled coil containing |
| chr14_-_28568107 | 0.06 |
ENSDART00000042850
ENSDART00000145502 |
insb
|
preproinsulin b |
| chr15_-_11341635 | 0.06 |
ENSDART00000055220
|
rab30
|
RAB30, member RAS oncogene family |
| chr10_+_7719796 | 0.06 |
ENSDART00000191795
|
ggcx
|
gamma-glutamyl carboxylase |
| chr2_-_57110477 | 0.06 |
ENSDART00000181132
|
slc25a42
|
solute carrier family 25, member 42 |
| chr19_+_9790806 | 0.06 |
ENSDART00000155948
|
cacng6a
|
calcium channel, voltage-dependent, gamma subunit 6a |
| chr5_+_15495351 | 0.06 |
ENSDART00000111646
ENSDART00000114446 |
suds3
|
SDS3 homolog, SIN3A corepressor complex component |
| chr15_-_12500938 | 0.06 |
ENSDART00000159627
|
scn4ba
|
sodium channel, voltage-gated, type IV, beta a |
| chr17_+_31914877 | 0.06 |
ENSDART00000177801
|
FAM196A (1 of many)
|
family with sequence similarity 196 member A |
| chr19_-_1023051 | 0.06 |
ENSDART00000158429
|
tmem42b
|
transmembrane protein 42b |
| chr11_+_33312601 | 0.06 |
ENSDART00000188024
|
cntnap5l
|
contactin associated protein-like 5 like |
| chr17_+_32571584 | 0.06 |
ENSDART00000087565
|
eva1a
|
eva-1 homolog A (C. elegans) |
| chr19_+_41701660 | 0.06 |
ENSDART00000033362
|
gatad2b
|
GATA zinc finger domain containing 2B |
| chr11_+_6739433 | 0.06 |
ENSDART00000163397
|
pde4cb
|
phosphodiesterase 4C, cAMP-specific b |
| chr18_+_5308392 | 0.06 |
ENSDART00000179072
|
DUT
|
deoxyuridine triphosphatase |
| chr19_-_12967986 | 0.05 |
ENSDART00000151064
|
slc25a32a
|
solute carrier family 25 (mitochondrial folate carrier), member 32a |
| chr18_-_41164277 | 0.05 |
ENSDART00000187766
ENSDART00000185375 |
CABZ01005876.1
|
|
| chr1_-_55196103 | 0.05 |
ENSDART00000140153
|
mri1
|
methylthioribose-1-phosphate isomerase 1 |
| chr2_-_12243213 | 0.05 |
ENSDART00000113081
|
gpr158b
|
G protein-coupled receptor 158b |
| chr15_-_18223769 | 0.05 |
ENSDART00000139966
|
or131-1
|
odorant receptor, family H, subfamily 131, member 1 |
| chr9_+_27876146 | 0.05 |
ENSDART00000133997
|
armc8
|
armadillo repeat containing 8 |
| chr6_-_50730749 | 0.05 |
ENSDART00000157153
ENSDART00000110441 |
pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr17_+_44463230 | 0.05 |
ENSDART00000130311
|
naa30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr15_-_766015 | 0.05 |
ENSDART00000190648
|
si:dkey-7i4.15
|
si:dkey-7i4.15 |
| chr6_+_3693441 | 0.05 |
ENSDART00000065256
|
ppig
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr25_-_20258508 | 0.04 |
ENSDART00000133860
ENSDART00000006840 ENSDART00000173434 |
dnm1l
|
dynamin 1-like |
| chr5_+_54685175 | 0.04 |
ENSDART00000115016
|
pmchl
|
pro-melanin-concentrating hormone, like |
| chr24_+_19591893 | 0.04 |
ENSDART00000152026
|
slco5a1a
|
solute carrier organic anion transporter family member 5A1a |
| chr3_-_50127668 | 0.04 |
ENSDART00000062116
|
cldnk
|
claudin k |
| chr23_+_3611765 | 0.04 |
ENSDART00000181481
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr9_+_29985010 | 0.04 |
ENSDART00000020743
|
cmss1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr23_-_24825863 | 0.04 |
ENSDART00000112493
|
syt6a
|
synaptotagmin VIa |
| chr22_+_696931 | 0.04 |
ENSDART00000149712
ENSDART00000009756 |
gpr37l1a
|
G protein-coupled receptor 37 like 1a |
| chr11_-_2222440 | 0.04 |
ENSDART00000162167
|
smug1
|
single-strand-selective monofunctional uracil-DNA glycosylase 1 |
| chr21_-_23046606 | 0.04 |
ENSDART00000016167
|
zw10
|
zw10 kinetochore protein |
| chr22_+_15747337 | 0.04 |
ENSDART00000062582
|
si:dkeyp-70f9.7
|
si:dkeyp-70f9.7 |
| chr18_+_33818577 | 0.04 |
ENSDART00000131788
|
olfcq19
|
olfactory receptor C family, q19 |
| chr19_+_48117995 | 0.04 |
ENSDART00000170865
|
nme2b.1
|
NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 1 |
| chr11_+_27134116 | 0.04 |
ENSDART00000129736
|
hdac11
|
histone deacetylase 11 |
| chr15_+_47161917 | 0.03 |
ENSDART00000167860
|
gap43
|
growth associated protein 43 |
| chr18_+_28102620 | 0.03 |
ENSDART00000132342
|
kiaa1549lb
|
KIAA1549-like b |
| chr6_+_53361874 | 0.03 |
ENSDART00000180660
|
si:ch211-161c3.5
|
si:ch211-161c3.5 |
| chr7_-_11605185 | 0.03 |
ENSDART00000169291
ENSDART00000113904 |
stard5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr9_-_27442339 | 0.03 |
ENSDART00000138602
|
stxbp5l
|
syntaxin binding protein 5-like |
| chr21_+_21791343 | 0.03 |
ENSDART00000151654
|
neu3.1
|
sialidase 3 (membrane sialidase), tandem duplicate 1 |
| chr16_-_39900665 | 0.03 |
ENSDART00000136719
|
rbms3
|
RNA binding motif, single stranded interacting protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of znf143b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.2 | 0.5 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.8 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 0.5 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.2 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.1 | 0.3 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 0.2 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.1 | 0.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.2 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.1 | 0.3 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.3 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.1 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.1 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.2 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.3 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.4 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.1 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.0 | 0.2 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:0030816 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.3 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.1 | GO:0017003 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.1 | 0.2 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.2 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.0 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.2 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.3 | GO:0015369 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.1 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.5 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.3 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) FAD transmembrane transporter activity(GO:0015230) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.0 | 0.1 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.0 | 0.0 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.0 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |