Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
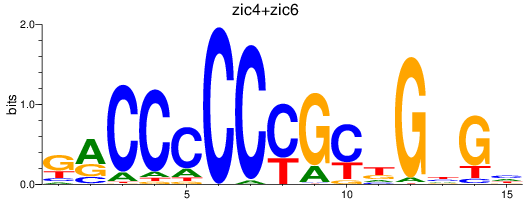
Results for zic4+zic6
Z-value: 0.28
Transcription factors associated with zic4+zic6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zic4
|
ENSDARG00000031307 | zic family member 4 |
|
zic6
|
ENSDARG00000071496 | zic family member 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zic4 | dr11_v1_chr24_+_4977862_4977862 | -0.81 | 4.3e-05 | Click! |
| zic6 | dr11_v1_chr14_+_32022272_32022272 | 0.09 | 7.3e-01 | Click! |
Activity profile of zic4+zic6 motif
Sorted Z-values of zic4+zic6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_4245311 | 1.22 |
ENSDART00000055379
|
gdf3
|
growth differentiation factor 3 |
| chr17_-_4245902 | 1.05 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr19_-_27550768 | 0.84 |
ENSDART00000142313
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr8_+_15269423 | 0.61 |
ENSDART00000020386
|
gclm
|
glutamate-cysteine ligase, modifier subunit |
| chr12_-_4243268 | 0.57 |
ENSDART00000131275
|
zgc:92313
|
zgc:92313 |
| chr15_+_47746176 | 0.46 |
ENSDART00000154481
ENSDART00000160914 |
stard10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr24_-_34680956 | 0.45 |
ENSDART00000171009
|
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr1_-_34450784 | 0.45 |
ENSDART00000140515
|
lmo7b
|
LIM domain 7b |
| chr3_+_24094581 | 0.45 |
ENSDART00000138270
ENSDART00000131509 |
copz2
|
coatomer protein complex, subunit zeta 2 |
| chr1_-_34450622 | 0.42 |
ENSDART00000083736
|
lmo7b
|
LIM domain 7b |
| chr5_-_15948833 | 0.40 |
ENSDART00000051649
ENSDART00000124467 |
xbp1
|
X-box binding protein 1 |
| chr5_+_31811662 | 0.37 |
ENSDART00000023463
|
uap1l1
|
UDP-N-acetylglucosamine pyrophosphorylase 1, like 1 |
| chr5_+_13521081 | 0.35 |
ENSDART00000171975
|
si:ch211-230g14.6
|
si:ch211-230g14.6 |
| chr12_-_41619257 | 0.33 |
ENSDART00000162967
|
dpysl4
|
dihydropyrimidinase-like 4 |
| chr1_+_40613297 | 0.33 |
ENSDART00000040798
ENSDART00000168067 ENSDART00000130490 |
naa15b
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit b |
| chr11_-_2504750 | 0.31 |
ENSDART00000173149
|
dgkab
|
diacylglycerol kinase, alpha b |
| chr19_-_81851 | 0.30 |
ENSDART00000172319
|
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr11_-_44999858 | 0.29 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr6_+_23026714 | 0.26 |
ENSDART00000124948
|
srp68
|
signal recognition particle 68 |
| chr19_-_17996162 | 0.26 |
ENSDART00000150928
ENSDART00000104491 |
ints8
|
integrator complex subunit 8 |
| chr5_+_37744625 | 0.26 |
ENSDART00000014031
|
dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr24_-_21172122 | 0.26 |
ENSDART00000154259
|
atp6v1ab
|
ATPase H+ transporting V1 subunit Ab |
| chr20_-_14781904 | 0.25 |
ENSDART00000187200
ENSDART00000179912 ENSDART00000160481 ENSDART00000026969 |
suco
|
SUN domain containing ossification factor |
| chr19_-_82504 | 0.24 |
ENSDART00000027864
ENSDART00000160560 |
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr18_+_31117136 | 0.24 |
ENSDART00000138403
|
tcf25
|
transcription factor 25 (basic helix-loop-helix) |
| chr8_-_4618653 | 0.23 |
ENSDART00000025535
|
sept5a
|
septin 5a |
| chr12_+_20627164 | 0.23 |
ENSDART00000190179
|
stx4
|
syntaxin 4 |
| chr12_-_2800809 | 0.23 |
ENSDART00000152682
ENSDART00000083784 |
ubtd1b
|
ubiquitin domain containing 1b |
| chr25_-_1079417 | 0.23 |
ENSDART00000163134
|
FO907089.1
|
|
| chr12_+_20627505 | 0.23 |
ENSDART00000074384
|
stx4
|
syntaxin 4 |
| chr18_+_1615 | 0.23 |
ENSDART00000082450
|
homer2
|
homer scaffolding protein 2 |
| chr8_-_1266181 | 0.23 |
ENSDART00000148654
ENSDART00000149924 |
cdc14b
|
cell division cycle 14B |
| chr19_-_17996336 | 0.22 |
ENSDART00000186143
ENSDART00000080751 |
ints8
|
integrator complex subunit 8 |
| chr17_-_23895026 | 0.22 |
ENSDART00000122108
|
pdzd8
|
PDZ domain containing 8 |
| chr18_-_20458412 | 0.21 |
ENSDART00000012241
|
kif23
|
kinesin family member 23 |
| chr13_+_37653851 | 0.21 |
ENSDART00000141988
ENSDART00000126902 ENSDART00000100352 |
phf3
|
PHD finger protein 3 |
| chr10_-_4961923 | 0.20 |
ENSDART00000050177
ENSDART00000146066 |
snx30
|
sorting nexin family member 30 |
| chr18_+_20047374 | 0.19 |
ENSDART00000146957
|
uacaa
|
uveal autoantigen with coiled-coil domains and ankyrin repeats a |
| chr5_+_36611128 | 0.17 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr21_-_217589 | 0.17 |
ENSDART00000185017
|
CZQB01146713.1
|
|
| chr21_+_5926777 | 0.16 |
ENSDART00000121769
|
rexo4
|
REX4 homolog, 3'-5' exonuclease |
| chr17_+_52300018 | 0.16 |
ENSDART00000190302
|
esrrb
|
estrogen-related receptor beta |
| chr1_-_58601636 | 0.16 |
ENSDART00000141143
|
si:ch73-236c18.8
|
si:ch73-236c18.8 |
| chr21_-_14664445 | 0.15 |
ENSDART00000124223
|
ehmt1b
|
euchromatic histone-lysine N-methyltransferase 1b |
| chr20_-_28768109 | 0.14 |
ENSDART00000114611
ENSDART00000182443 |
sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr21_+_3093419 | 0.14 |
ENSDART00000162520
|
SHC3
|
SHC adaptor protein 3 |
| chr19_+_40122160 | 0.14 |
ENSDART00000143966
|
si:ch211-173p18.3
|
si:ch211-173p18.3 |
| chr13_+_16521898 | 0.14 |
ENSDART00000122557
|
kcnma1a
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1a |
| chr25_+_2776511 | 0.14 |
ENSDART00000115280
|
neo1b
|
neogenin 1b |
| chr21_-_21465111 | 0.13 |
ENSDART00000141487
|
nectin3b
|
nectin cell adhesion molecule 3b |
| chr11_+_34921492 | 0.13 |
ENSDART00000128070
|
gnai2a
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2a |
| chr8_+_26859639 | 0.11 |
ENSDART00000133440
|
prdm2a
|
PR domain containing 2, with ZNF domain a |
| chr22_-_26558166 | 0.11 |
ENSDART00000111125
|
glis2a
|
GLIS family zinc finger 2a |
| chr5_+_30596822 | 0.10 |
ENSDART00000188375
|
hinfp
|
histone H4 transcription factor |
| chr11_+_31558006 | 0.10 |
ENSDART00000024296
|
egln1b
|
egl-9 family hypoxia-inducible factor 1b |
| chr24_-_20599781 | 0.09 |
ENSDART00000179664
ENSDART00000141823 |
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr16_-_7228276 | 0.09 |
ENSDART00000149030
|
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr3_-_40162843 | 0.08 |
ENSDART00000129664
ENSDART00000025285 |
drg2
|
developmentally regulated GTP binding protein 2 |
| chr14_+_40852497 | 0.07 |
ENSDART00000128588
ENSDART00000166065 |
taf7
|
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr25_-_27729046 | 0.07 |
ENSDART00000131437
|
zgc:153935
|
zgc:153935 |
| chr15_-_33495048 | 0.07 |
ENSDART00000159882
|
stard13b
|
StAR-related lipid transfer (START) domain containing 13b |
| chr5_-_67750907 | 0.07 |
ENSDART00000172097
|
b4galt4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr20_-_28642061 | 0.07 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr3_-_19561058 | 0.06 |
ENSDART00000079323
|
zgc:163079
|
zgc:163079 |
| chr23_-_27547931 | 0.06 |
ENSDART00000144419
|
larp4aa
|
La ribonucleoprotein domain family, member 4Aa |
| chr19_+_9790806 | 0.06 |
ENSDART00000155948
|
cacng6a
|
calcium channel, voltage-dependent, gamma subunit 6a |
| chr17_+_34215886 | 0.06 |
ENSDART00000186775
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr2_-_42628028 | 0.05 |
ENSDART00000179866
|
myo10
|
myosin X |
| chr15_-_26844591 | 0.05 |
ENSDART00000077582
|
pitpnm3
|
PITPNM family member 3 |
| chr24_-_10897511 | 0.05 |
ENSDART00000145593
ENSDART00000102484 ENSDART00000066784 |
fam49bb
|
family with sequence similarity 49, member Bb |
| chr5_-_68927728 | 0.05 |
ENSDART00000132838
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr3_-_37588855 | 0.05 |
ENSDART00000149258
|
arf2a
|
ADP-ribosylation factor 2a |
| chr14_-_21097574 | 0.04 |
ENSDART00000186803
|
rnf20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr24_-_21587335 | 0.04 |
ENSDART00000091528
|
gpr12
|
G protein-coupled receptor 12 |
| chr11_+_31558207 | 0.04 |
ENSDART00000140204
|
egln1b
|
egl-9 family hypoxia-inducible factor 1b |
| chr22_-_18116635 | 0.04 |
ENSDART00000005724
|
ncanb
|
neurocan b |
| chr4_+_76349919 | 0.04 |
ENSDART00000174194
|
si:ch73-158p21.2
|
si:ch73-158p21.2 |
| chr1_-_54191626 | 0.04 |
ENSDART00000062941
|
nthl1
|
nth-like DNA glycosylase 1 |
| chr5_-_23725480 | 0.04 |
ENSDART00000137063
|
si:dkey-110k5.10
|
si:dkey-110k5.10 |
| chr17_+_28670132 | 0.04 |
ENSDART00000076344
ENSDART00000164981 ENSDART00000182851 |
hectd1
|
HECT domain containing 1 |
| chr5_+_31944270 | 0.03 |
ENSDART00000147850
|
ungb
|
uracil DNA glycosylase b |
| chr15_-_38202630 | 0.03 |
ENSDART00000183772
|
rhoga
|
ras homolog family member Ga |
| chr12_-_4206869 | 0.03 |
ENSDART00000106572
|
si:dkey-32n7.9
|
si:dkey-32n7.9 |
| chr17_-_14523722 | 0.03 |
ENSDART00000024726
|
daam1a
|
dishevelled associated activator of morphogenesis 1a |
| chr24_-_13566015 | 0.03 |
ENSDART00000123450
|
KCNB2
|
si:dkey-192j17.1 |
| chr2_-_14987282 | 0.03 |
ENSDART00000143057
|
hccsa.2
|
holocytochrome c synthase a, tandem duplicate 2 |
| chr16_-_55028740 | 0.02 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr5_-_61588998 | 0.02 |
ENSDART00000050912
|
pex12
|
peroxisomal biogenesis factor 12 |
| chr21_-_33995213 | 0.02 |
ENSDART00000140184
|
EBF1 (1 of many)
|
si:ch211-51e8.2 |
| chr18_-_50947868 | 0.02 |
ENSDART00000174276
|
st7
|
suppression of tumorigenicity 7 |
| chr8_+_40210398 | 0.02 |
ENSDART00000167612
|
rnf34a
|
ring finger protein 34a |
| chr17_+_132555 | 0.02 |
ENSDART00000158159
|
zgc:77287
|
zgc:77287 |
| chr10_+_10677697 | 0.01 |
ENSDART00000188705
|
fam163b
|
family with sequence similarity 163, member B |
| chr16_-_46567344 | 0.01 |
ENSDART00000127721
|
si:dkey-152b24.7
|
si:dkey-152b24.7 |
| chr25_-_18730697 | 0.01 |
ENSDART00000182475
|
si:dkeyp-93a5.2
|
si:dkeyp-93a5.2 |
| chr6_-_7438584 | 0.01 |
ENSDART00000053776
|
fkbp11
|
FK506 binding protein 11 |
| chr9_-_32191620 | 0.01 |
ENSDART00000139426
|
pimr143
|
Pim proto-oncogene, serine/threonine kinase, related 143 |
| chr4_+_64562090 | 0.01 |
ENSDART00000188810
|
si:ch211-223a21.3
|
si:ch211-223a21.3 |
| chr10_+_21786656 | 0.01 |
ENSDART00000185851
ENSDART00000167219 |
pcdh1g26
|
protocadherin 1 gamma 26 |
| chr9_-_37748513 | 0.01 |
ENSDART00000188967
ENSDART00000187886 |
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr23_-_29824146 | 0.00 |
ENSDART00000020616
|
zgc:194189
|
zgc:194189 |
| chr3_+_46724528 | 0.00 |
ENSDART00000181358
|
pde4a
|
phosphodiesterase 4A, cAMP-specific |
| chr18_-_15559817 | 0.00 |
ENSDART00000061681
|
si:ch211-245j22.3
|
si:ch211-245j22.3 |
| chr3_-_19899914 | 0.00 |
ENSDART00000134969
|
rnd2
|
Rho family GTPase 2 |
| chr13_-_32648382 | 0.00 |
ENSDART00000138571
ENSDART00000132820 |
bend3
|
BEN domain containing 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of zic4+zic6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.3 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.2 | 0.6 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 0.3 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.1 | 0.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.2 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.0 | 0.4 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.2 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.0 | 0.2 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.2 | GO:0006203 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.0 | 0.3 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.5 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.5 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.1 | GO:0035587 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.1 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.0 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 0.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0008832 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.0 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 2.1 | GO:0008083 | growth factor activity(GO:0008083) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 2.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |