Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
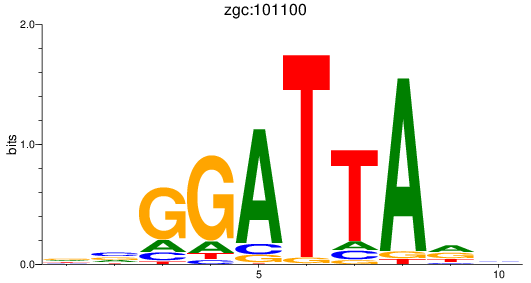
Results for zgc:101100
Z-value: 0.29
Transcription factors associated with zgc:101100
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zgc
|
ENSDARG00000021959 | 101100 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zgc:101100 | dr11_v1_chr8_+_20488322_20488322 | 0.82 | 3.3e-05 | Click! |
Activity profile of zgc:101100 motif
Sorted Z-values of zgc:101100 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_2841205 | 0.70 |
ENSDART00000131505
ENSDART00000055869 |
ykt6
|
YKT6 v-SNARE homolog (S. cerevisiae) |
| chr8_+_2656231 | 0.58 |
ENSDART00000160833
|
fam102aa
|
family with sequence similarity 102, member Aa |
| chr1_-_44928987 | 0.53 |
ENSDART00000134635
|
si:dkey-9i23.15
|
si:dkey-9i23.15 |
| chr8_+_2656681 | 0.52 |
ENSDART00000185067
ENSDART00000165943 |
fam102aa
|
family with sequence similarity 102, member Aa |
| chr21_-_9562272 | 0.49 |
ENSDART00000162225
ENSDART00000163874 ENSDART00000168173 |
ptpn13
|
protein tyrosine phosphatase, non-receptor type 13 |
| chr7_+_66884291 | 0.48 |
ENSDART00000187499
|
sbf2
|
SET binding factor 2 |
| chr2_-_58075414 | 0.46 |
ENSDART00000161920
|
nectin4
|
nectin cell adhesion molecule 4 |
| chr6_-_17999776 | 0.46 |
ENSDART00000183048
ENSDART00000181577 ENSDART00000170597 |
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr22_-_20695237 | 0.46 |
ENSDART00000112722
|
org
|
oogenesis-related gene |
| chr18_+_49969568 | 0.46 |
ENSDART00000126916
|
mob2b
|
MOB kinase activator 2b |
| chr16_+_16265850 | 0.45 |
ENSDART00000181265
|
setd2
|
SET domain containing 2 |
| chr3_-_23513155 | 0.44 |
ENSDART00000170200
|
BX682558.1
|
|
| chr19_+_24068223 | 0.41 |
ENSDART00000141351
ENSDART00000100420 |
pex11b
|
peroxisomal biogenesis factor 11 beta |
| chr5_-_24029228 | 0.41 |
ENSDART00000051546
|
rps6ka3a
|
ribosomal protein S6 kinase a, polypeptide 3a |
| chr7_+_66884570 | 0.39 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr8_-_11016766 | 0.37 |
ENSDART00000064066
|
bcas2
|
BCAS2, pre-mRNA processing factor |
| chr20_-_25643667 | 0.35 |
ENSDART00000137457
|
si:dkeyp-117h8.4
|
si:dkeyp-117h8.4 |
| chr9_-_34500197 | 0.34 |
ENSDART00000114043
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr13_-_36525982 | 0.33 |
ENSDART00000114744
|
pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr8_-_25814263 | 0.31 |
ENSDART00000143397
|
taf10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr9_-_28275600 | 0.31 |
ENSDART00000170094
|
creb1b
|
cAMP responsive element binding protein 1b |
| chr17_+_35097024 | 0.29 |
ENSDART00000026152
|
asap2a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2a |
| chr20_+_34671386 | 0.27 |
ENSDART00000152836
ENSDART00000138226 |
elp3
|
elongator acetyltransferase complex subunit 3 |
| chr2_-_9696283 | 0.26 |
ENSDART00000165712
|
selenot1a
|
selenoprotein T, 1a |
| chr24_-_25004553 | 0.26 |
ENSDART00000080997
ENSDART00000136860 |
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr11_+_31680513 | 0.26 |
ENSDART00000139900
ENSDART00000040305 |
diaph3
|
diaphanous-related formin 3 |
| chr15_-_26887028 | 0.25 |
ENSDART00000156292
|
si:dkey-243i1.1
|
si:dkey-243i1.1 |
| chr22_+_438714 | 0.23 |
ENSDART00000136491
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr22_+_34701848 | 0.22 |
ENSDART00000082066
|
atpv0e2
|
ATPase H+ transporting V0 subunit e2 |
| chr2_-_30784198 | 0.21 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
| chr2_+_30531726 | 0.21 |
ENSDART00000146518
|
ankrd33bb
|
ankyrin repeat domain 33Bb |
| chr21_+_44773237 | 0.18 |
ENSDART00000084780
|
rnf121
|
ring finger protein 121 |
| chr16_+_11297703 | 0.18 |
ENSDART00000125158
|
znf574
|
zinc finger protein 574 |
| chr7_-_57933736 | 0.17 |
ENSDART00000142580
|
ank2b
|
ankyrin 2b, neuronal |
| chr7_-_49594995 | 0.16 |
ENSDART00000174161
ENSDART00000109147 |
brsk2b
|
BR serine/threonine kinase 2b |
| chr5_+_31791001 | 0.16 |
ENSDART00000043010
|
slc25a25b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25b |
| chr18_-_38245062 | 0.15 |
ENSDART00000189092
|
nat10
|
N-acetyltransferase 10 |
| chr25_-_32381642 | 0.15 |
ENSDART00000133872
ENSDART00000006124 |
cops2
|
COP9 signalosome subunit 2 |
| chr14_+_30753666 | 0.15 |
ENSDART00000009385
ENSDART00000137782 ENSDART00000172778 |
yif1a
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr24_+_37533728 | 0.14 |
ENSDART00000061203
|
rhot2
|
ras homolog family member T2 |
| chr12_+_33460794 | 0.14 |
ENSDART00000007053
ENSDART00000142716 |
narf
|
nuclear prelamin A recognition factor |
| chr5_+_47882319 | 0.14 |
ENSDART00000149316
|
rasa1a
|
RAS p21 protein activator (GTPase activating protein) 1a |
| chr21_-_41369370 | 0.13 |
ENSDART00000159290
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr10_+_6013076 | 0.12 |
ENSDART00000167613
ENSDART00000159216 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr6_-_59381391 | 0.12 |
ENSDART00000157066
|
ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr2_+_30787128 | 0.12 |
ENSDART00000189233
|
atp6v1h
|
ATPase H+ transporting V1 subunit H |
| chr14_+_32838110 | 0.12 |
ENSDART00000158077
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr3_-_10970502 | 0.12 |
ENSDART00000127500
|
CR382337.1
|
|
| chr2_+_30786773 | 0.12 |
ENSDART00000019029
ENSDART00000145681 |
atp6v1h
|
ATPase H+ transporting V1 subunit H |
| chr5_-_38107741 | 0.11 |
ENSDART00000156853
|
si:ch211-284e13.14
|
si:ch211-284e13.14 |
| chr6_+_13083146 | 0.11 |
ENSDART00000172158
|
l3hypdh
|
trans-L-3-hydroxyproline dehydratase |
| chr23_-_10137322 | 0.10 |
ENSDART00000142442
|
plxnb1a
|
plexin b1a |
| chr11_+_25583950 | 0.10 |
ENSDART00000111961
|
ccdc120
|
coiled-coil domain containing 120 |
| chr24_-_7777389 | 0.10 |
ENSDART00000138541
|
rpgrip1
|
RPGR interacting protein 1 |
| chr19_+_2631565 | 0.10 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr1_+_41690402 | 0.10 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr10_+_17026870 | 0.09 |
ENSDART00000184529
ENSDART00000157480 |
CR855996.2
|
|
| chr13_+_1015749 | 0.09 |
ENSDART00000190982
|
prokr1b
|
prokineticin receptor 1b |
| chr15_+_42995381 | 0.08 |
ENSDART00000152222
|
acsl3a
|
acyl-CoA synthetase long chain family member 3a |
| chr14_+_32837914 | 0.08 |
ENSDART00000158888
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr8_-_14121634 | 0.07 |
ENSDART00000184946
|
bgna
|
biglycan a |
| chr2_+_4146299 | 0.06 |
ENSDART00000173418
|
mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr18_-_25646286 | 0.06 |
ENSDART00000099511
ENSDART00000186890 |
si:ch211-13k12.2
|
si:ch211-13k12.2 |
| chr17_-_50010121 | 0.05 |
ENSDART00000122747
|
tmem30aa
|
transmembrane protein 30Aa |
| chr6_-_33924883 | 0.04 |
ENSDART00000132762
ENSDART00000148142 ENSDART00000142213 |
akr1a1b
|
aldo-keto reductase family 1, member A1b (aldehyde reductase) |
| chr19_-_40906161 | 0.04 |
ENSDART00000004514
|
bet1
|
Bet1 golgi vesicular membrane trafficking protein |
| chr22_+_35275468 | 0.04 |
ENSDART00000189516
ENSDART00000181572 ENSDART00000165353 ENSDART00000185352 |
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr11_+_37909654 | 0.04 |
ENSDART00000172211
|
si:ch211-112f3.4
|
si:ch211-112f3.4 |
| chr21_-_2348838 | 0.03 |
ENSDART00000160337
|
si:ch73-299h12.8
|
si:ch73-299h12.8 |
| chr18_+_7639401 | 0.03 |
ENSDART00000092416
|
rabl2
|
RAB, member of RAS oncogene family-like 2 |
| chr2_+_4146606 | 0.03 |
ENSDART00000171170
|
mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr25_-_17503393 | 0.03 |
ENSDART00000061726
|
trim35-40
|
tripartite motif containing 35-40 |
| chr8_-_54077740 | 0.03 |
ENSDART00000027000
|
rho
|
rhodopsin |
| chr23_-_18981595 | 0.03 |
ENSDART00000147617
|
bcl2l1
|
bcl2-like 1 |
| chr1_-_45146834 | 0.02 |
ENSDART00000144997
|
si:ch211-239f4.6
|
si:ch211-239f4.6 |
| chr12_+_6195191 | 0.02 |
ENSDART00000043236
ENSDART00000186420 |
prkg1b
|
protein kinase, cGMP-dependent, type Ib |
| chr11_-_15090118 | 0.02 |
ENSDART00000171118
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr17_-_2690083 | 0.02 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr17_-_51202339 | 0.01 |
ENSDART00000167117
|
si:ch1073-469d17.2
|
si:ch1073-469d17.2 |
| chr19_+_4061699 | 0.01 |
ENSDART00000158309
ENSDART00000166512 |
btr25
btr26
|
bloodthirsty-related gene family, member 25 bloodthirsty-related gene family, member 26 |
| chr7_-_17780048 | 0.01 |
ENSDART00000183336
|
si:dkey-106g10.7
|
si:dkey-106g10.7 |
| chr20_-_18736281 | 0.00 |
ENSDART00000142837
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr9_+_21990095 | 0.00 |
ENSDART00000146829
ENSDART00000133515 ENSDART00000193582 |
si:dkey-57a22.13
|
si:dkey-57a22.13 |
| chr11_+_30296332 | 0.00 |
ENSDART00000192843
|
ugt1b7
|
UDP glucuronosyltransferase 1 family, polypeptide B7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of zgc:101100
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.3 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.1 | 0.5 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.1 | 0.4 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 0.2 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.0 | 0.1 | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.0 | 0.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.2 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.1 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.5 | GO:0038202 | TORC1 signaling(GO:0038202) cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 0.0 | GO:0090219 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.0 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.4 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.4 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.0 | 0.5 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.5 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.2 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.0 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.2 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |