Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
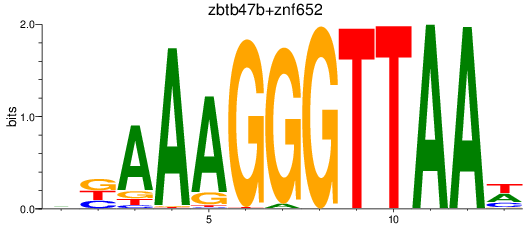
Results for zbtb47b+znf652
Z-value: 1.01
Transcription factors associated with zbtb47b+znf652
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
znf652
|
ENSDARG00000062302 | zinc finger protein 652 |
|
zbtb47b
|
ENSDARG00000079547 | zinc finger and BTB domain containing 47b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zbtb47b | dr11_v1_chr24_-_20641000_20641000 | -0.96 | 4.2e-10 | Click! |
| znf652 | dr11_v1_chr3_+_15773991_15773991 | -0.92 | 5.8e-08 | Click! |
Activity profile of zbtb47b+znf652 motif
Sorted Z-values of zbtb47b+znf652 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_38643188 | 2.94 |
ENSDART00000144972
|
si:ch211-271e10.1
|
si:ch211-271e10.1 |
| chr20_-_52631998 | 2.76 |
ENSDART00000145283
ENSDART00000139072 |
si:ch211-221n20.1
|
si:ch211-221n20.1 |
| chr14_+_45565891 | 2.46 |
ENSDART00000133389
ENSDART00000025549 |
zgc:92249
|
zgc:92249 |
| chr11_-_3959889 | 2.45 |
ENSDART00000159683
|
pbrm1
|
polybromo 1 |
| chr16_-_29557338 | 2.41 |
ENSDART00000058888
|
hormad1
|
HORMA domain containing 1 |
| chr5_+_37978501 | 2.38 |
ENSDART00000012050
|
apoa1a
|
apolipoprotein A-Ia |
| chr10_+_44042033 | 2.10 |
ENSDART00000190006
ENSDART00000046172 |
cryba4
|
crystallin, beta A4 |
| chr7_-_19544575 | 2.07 |
ENSDART00000161549
|
CCNB1IP1
|
si:ch211-212k18.11 |
| chr20_-_52666150 | 1.99 |
ENSDART00000146716
ENSDART00000134155 |
si:ch211-221n20.4
|
si:ch211-221n20.4 |
| chr6_-_2627488 | 1.99 |
ENSDART00000044089
ENSDART00000158333 ENSDART00000155109 |
hyi
|
hydroxypyruvate isomerase |
| chr5_-_30535327 | 1.96 |
ENSDART00000040328
|
h2afx
|
H2A histone family, member X |
| chr5_-_10012971 | 1.83 |
ENSDART00000179421
|
CR936408.2
|
|
| chr7_+_20109968 | 1.79 |
ENSDART00000146335
|
zgc:114045
|
zgc:114045 |
| chr8_-_51579286 | 1.77 |
ENSDART00000147878
|
ankrd39
|
ankyrin repeat domain 39 |
| chr10_-_29744921 | 1.72 |
ENSDART00000088605
|
ift46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr11_+_2633743 | 1.70 |
ENSDART00000173298
|
si:ch211-160o17.3
|
si:ch211-160o17.3 |
| chr23_+_2542158 | 1.66 |
ENSDART00000182197
|
LO017835.1
|
|
| chr2_+_44571200 | 1.65 |
ENSDART00000098132
|
klhl24a
|
kelch-like family member 24a |
| chr8_-_51578926 | 1.64 |
ENSDART00000190625
|
ankrd39
|
ankyrin repeat domain 39 |
| chr11_+_3959495 | 1.62 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr7_-_52963493 | 1.56 |
ENSDART00000052029
|
cart3
|
cocaine- and amphetamine-regulated transcript 3 |
| chr19_+_13420884 | 1.56 |
ENSDART00000160209
|
si:ch211-204a13.2
|
si:ch211-204a13.2 |
| chr7_-_40110140 | 1.52 |
ENSDART00000173469
|
si:ch73-174h16.5
|
si:ch73-174h16.5 |
| chr14_+_17397534 | 1.52 |
ENSDART00000180162
ENSDART00000129838 |
rnf212
|
ring finger protein 212 |
| chr7_-_7823662 | 1.51 |
ENSDART00000167652
|
cxcl8b.3
|
chemokine (C-X-C motif) ligand 8b, duplicate 3 |
| chr17_+_40989973 | 1.49 |
ENSDART00000160049
|
mrpl33
|
mitochondrial ribosomal protein L33 |
| chr24_+_40860320 | 1.49 |
ENSDART00000161351
|
gorasp1b
|
golgi reassembly stacking protein 1b |
| chr4_-_8611841 | 1.48 |
ENSDART00000067322
|
fbxl14b
|
F-box and leucine-rich repeat protein 14b |
| chr8_+_39760258 | 1.48 |
ENSDART00000037914
|
cox6a1
|
cytochrome c oxidase subunit VIa polypeptide 1 |
| chr11_-_3959477 | 1.47 |
ENSDART00000045971
|
pbrm1
|
polybromo 1 |
| chr21_+_22423286 | 1.45 |
ENSDART00000133190
|
capslb
|
calcyphosine-like b |
| chr4_-_16545085 | 1.42 |
ENSDART00000033188
|
btg1
|
B-cell translocation gene 1, anti-proliferative |
| chr1_+_40291054 | 1.41 |
ENSDART00000136916
|
vwa10.1
|
von Willebrand factor A domain containing 10, tandem duplicate 1 |
| chr14_-_48939560 | 1.41 |
ENSDART00000021736
|
scocb
|
short coiled-coil protein b |
| chr17_-_6600899 | 1.41 |
ENSDART00000154074
ENSDART00000180912 |
ANKRD66
|
si:ch211-189e2.2 |
| chr13_+_36764715 | 1.38 |
ENSDART00000111832
ENSDART00000085230 |
atl1
|
atlastin GTPase 1 |
| chr2_-_33455164 | 1.36 |
ENSDART00000134024
ENSDART00000132221 |
ccdc24
|
coiled-coil domain containing 24 |
| chr19_+_43297546 | 1.35 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr16_-_40508624 | 1.34 |
ENSDART00000075718
|
ndufaf6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr13_+_502230 | 1.34 |
ENSDART00000013007
|
degs1
|
delta(4)-desaturase, sphingolipid 1 |
| chr16_-_42056137 | 1.34 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr23_-_45339439 | 1.33 |
ENSDART00000148726
|
ccdc171
|
coiled-coil domain containing 171 |
| chr4_-_18595525 | 1.33 |
ENSDART00000049061
|
cdkn1ba
|
cyclin-dependent kinase inhibitor 1Ba |
| chr20_-_46534179 | 1.32 |
ENSDART00000060689
|
eif2b2
|
eukaryotic translation initiation factor 2B, subunit 2 beta |
| chr12_+_25097754 | 1.30 |
ENSDART00000050070
|
pigf
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr5_+_4298636 | 1.29 |
ENSDART00000100061
|
prdx4
|
peroxiredoxin 4 |
| chr1_-_45752010 | 1.26 |
ENSDART00000101410
|
si:ch211-214c7.5
|
si:ch211-214c7.5 |
| chr10_+_4987766 | 1.25 |
ENSDART00000121959
|
si:ch73-234b20.5
|
si:ch73-234b20.5 |
| chr14_-_49063157 | 1.24 |
ENSDART00000021260
|
sept8b
|
septin 8b |
| chr22_+_17237219 | 1.23 |
ENSDART00000090069
|
axdnd1
|
axonemal dynein light chain domain containing 1 |
| chr1_-_53714885 | 1.20 |
ENSDART00000026409
|
cct4
|
chaperonin containing TCP1, subunit 4 (delta) |
| chr14_-_215051 | 1.16 |
ENSDART00000054822
|
nkx3.2
|
NK3 homeobox 2 |
| chr23_+_25893020 | 1.14 |
ENSDART00000144769
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr1_-_45752261 | 1.11 |
ENSDART00000141756
|
si:ch211-214c7.5
|
si:ch211-214c7.5 |
| chr17_-_2535682 | 1.07 |
ENSDART00000155227
|
ccdc9b
|
coiled-coil domain containing 9B |
| chr13_+_47007075 | 1.04 |
ENSDART00000109247
ENSDART00000183205 ENSDART00000180924 ENSDART00000133146 |
anapc1
|
anaphase promoting complex subunit 1 |
| chr20_+_42761881 | 1.03 |
ENSDART00000113625
|
pimr113
|
Pim proto-oncogene, serine/threonine kinase, related 113 |
| chr24_-_23758003 | 1.02 |
ENSDART00000178085
|
BX640462.2
|
Danio rerio minichromosome maintenance domain containing 2 (mcmdc2), mRNA. |
| chr10_-_20357013 | 1.02 |
ENSDART00000080143
|
sfrp1b
|
secreted frizzled-related protein 1b |
| chr15_-_29162193 | 1.02 |
ENSDART00000138449
ENSDART00000099885 |
xaf1
|
XIAP associated factor 1 |
| chr17_+_26815021 | 1.01 |
ENSDART00000086885
|
asmt2
|
acetylserotonin O-methyltransferase 2 |
| chr6_+_43015916 | 1.00 |
ENSDART00000064888
|
tcta
|
T cell leukemia translocation altered |
| chr20_+_42780162 | 1.00 |
ENSDART00000127069
|
pimr213
|
Pim proto-oncogene, serine/threonine kinase, related 213 |
| chr7_+_58699718 | 0.99 |
ENSDART00000049264
|
sdr16c5b
|
short chain dehydrogenase/reductase family 16C, member 5b |
| chr16_+_10346277 | 0.97 |
ENSDART00000081092
|
si:dkeyp-77h1.4
|
si:dkeyp-77h1.4 |
| chr18_-_977075 | 0.97 |
ENSDART00000032392
|
dhdhl
|
dihydrodiol dehydrogenase (dimeric), like |
| chr8_-_13442871 | 0.96 |
ENSDART00000144887
|
pimr106
|
Pim proto-oncogene, serine/threonine kinase, related 106 |
| chr20_-_19365875 | 0.96 |
ENSDART00000063703
ENSDART00000187707 ENSDART00000161065 |
si:dkey-71h2.2
|
si:dkey-71h2.2 |
| chr20_+_52584532 | 0.95 |
ENSDART00000138641
|
si:dkey-235d18.5
|
si:dkey-235d18.5 |
| chr8_-_13428740 | 0.95 |
ENSDART00000131826
|
CR354547.3
|
|
| chr5_-_4418555 | 0.94 |
ENSDART00000170158
|
apooa
|
apolipoprotein O, a |
| chr7_-_29227786 | 0.92 |
ENSDART00000171300
|
herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr8_-_13471916 | 0.92 |
ENSDART00000146558
|
pimr105
|
Pim proto-oncogene, serine/threonine kinase, related 105 |
| chr3_-_16411244 | 0.91 |
ENSDART00000140067
|
eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr16_+_23913943 | 0.90 |
ENSDART00000175404
ENSDART00000129525 |
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr3_-_34027178 | 0.89 |
ENSDART00000170201
ENSDART00000151408 |
ighv1-4
ighv14-1
|
immunoglobulin heavy variable 1-4 immunoglobulin heavy variable 14-1 |
| chr20_-_43572763 | 0.88 |
ENSDART00000153251
|
pimr125
|
Pim proto-oncogene, serine/threonine kinase, related 125 |
| chr3_+_32112004 | 0.88 |
ENSDART00000105272
|
zgc:173593
|
zgc:173593 |
| chr16_+_9053275 | 0.88 |
ENSDART00000123150
|
dnah5
|
dynein, axonemal, heavy chain 5 |
| chr7_-_40993456 | 0.88 |
ENSDART00000031700
|
en2a
|
engrailed homeobox 2a |
| chr3_+_24207243 | 0.87 |
ENSDART00000023454
ENSDART00000136400 |
adsl
|
adenylosuccinate lyase |
| chr1_-_46505310 | 0.85 |
ENSDART00000178072
|
si:busm1-105l16.2
|
si:busm1-105l16.2 |
| chr4_-_56068511 | 0.85 |
ENSDART00000168345
|
znf1133
|
zinc finger protein 1133 |
| chr2_+_24931677 | 0.84 |
ENSDART00000021528
|
agtr1a
|
angiotensin II receptor, type 1a |
| chr4_-_119689 | 0.83 |
ENSDART00000161055
|
dusp16
|
dual specificity phosphatase 16 |
| chr23_+_23020709 | 0.83 |
ENSDART00000146463
|
samd11
|
sterile alpha motif domain containing 11 |
| chr14_-_17563773 | 0.81 |
ENSDART00000082667
|
fgfrl1a
|
fibroblast growth factor receptor like 1a |
| chr4_-_49952636 | 0.81 |
ENSDART00000157941
|
si:dkey-156k2.3
|
si:dkey-156k2.3 |
| chr24_+_9372292 | 0.81 |
ENSDART00000082422
ENSDART00000191127 ENSDART00000180510 |
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr14_-_83154 | 0.80 |
ENSDART00000187097
|
CABZ01086812.1
|
|
| chr21_+_45733871 | 0.80 |
ENSDART00000187285
ENSDART00000193018 |
zgc:77058
|
zgc:77058 |
| chr8_+_50531709 | 0.80 |
ENSDART00000193352
|
PEBP4
|
phosphatidylethanolamine binding protein 4 |
| chr8_-_13486258 | 0.80 |
ENSDART00000137459
|
pimr104
|
Pim proto-oncogene, serine/threonine kinase, related 104 |
| chr18_+_20567542 | 0.79 |
ENSDART00000182585
|
bida
|
BH3 interacting domain death agonist |
| chr18_-_17075098 | 0.77 |
ENSDART00000042496
ENSDART00000192284 ENSDART00000180307 |
tango6
|
transport and golgi organization 6 homolog (Drosophila) |
| chr5_+_57743815 | 0.77 |
ENSDART00000005090
|
alg9
|
ALG9, alpha-1,2-mannosyltransferase |
| chr18_+_16744307 | 0.76 |
ENSDART00000179872
ENSDART00000133490 |
lyve1b
|
lymphatic vessel endothelial hyaluronic receptor 1b |
| chr17_+_7510220 | 0.75 |
ENSDART00000192648
|
klhl10b.1
|
kelch-like family member 10b, tandem duplicate 1 |
| chr17_+_7510998 | 0.75 |
ENSDART00000182626
|
klhl10b.1
|
kelch-like family member 10b, tandem duplicate 1 |
| chr19_-_19339285 | 0.74 |
ENSDART00000158413
ENSDART00000170479 |
cspg5b
|
chondroitin sulfate proteoglycan 5b |
| chr6_+_9427641 | 0.74 |
ENSDART00000022620
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr11_-_45138857 | 0.74 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr20_+_51478939 | 0.73 |
ENSDART00000149758
|
tlr5a
|
toll-like receptor 5a |
| chr4_-_57530817 | 0.72 |
ENSDART00000158435
|
zgc:173702
|
zgc:173702 |
| chr8_+_13694950 | 0.72 |
ENSDART00000132506
|
pimr107
|
Pim proto-oncogene, serine/threonine kinase, related 107 |
| chr17_-_6536305 | 0.70 |
ENSDART00000154855
|
cenpo
|
centromere protein O |
| chr7_-_8738827 | 0.70 |
ENSDART00000172807
ENSDART00000173026 |
si:ch211-1o7.3
|
si:ch211-1o7.3 |
| chr12_-_48671612 | 0.69 |
ENSDART00000007202
|
zgc:92749
|
zgc:92749 |
| chr3_-_15859397 | 0.67 |
ENSDART00000135924
|
dhrs7b
|
dehydrogenase/reductase (SDR family) member 7B |
| chr8_-_13419049 | 0.67 |
ENSDART00000133656
|
pimr101
|
Pim proto-oncogene, serine/threonine kinase, related 101 |
| chr13_+_844150 | 0.67 |
ENSDART00000058260
|
gsta.1
|
glutathione S-transferase, alpha tandem duplicate 1 |
| chr18_-_37407235 | 0.67 |
ENSDART00000132315
|
cep126
|
centrosomal protein 126 |
| chr11_-_3343463 | 0.66 |
ENSDART00000066177
|
tuba2
|
tubulin, alpha 2 |
| chr5_+_9428876 | 0.66 |
ENSDART00000081791
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr8_+_2487883 | 0.66 |
ENSDART00000101841
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr17_-_53353653 | 0.66 |
ENSDART00000180744
ENSDART00000026879 |
unm_sa911
|
un-named sa911 |
| chr12_-_31461412 | 0.65 |
ENSDART00000175929
ENSDART00000186342 |
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr20_+_47143900 | 0.65 |
ENSDART00000153360
|
si:dkeyp-104f11.6
|
si:dkeyp-104f11.6 |
| chr22_+_14051245 | 0.64 |
ENSDART00000043711
ENSDART00000164259 |
aox6
|
aldehyde oxidase 6 |
| chr6_-_40697585 | 0.64 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr16_-_5115993 | 0.63 |
ENSDART00000138654
|
ttk
|
ttk protein kinase |
| chr7_+_58699900 | 0.63 |
ENSDART00000144009
|
sdr16c5b
|
short chain dehydrogenase/reductase family 16C, member 5b |
| chr8_-_13454281 | 0.61 |
ENSDART00000141959
|
CR354547.2
|
|
| chr9_+_38446504 | 0.61 |
ENSDART00000077479
|
cyp27a1.4
|
cytochrome P450, family 27, subfamily A, polypeptide 1, gene 4 |
| chr4_+_58576146 | 0.61 |
ENSDART00000164911
|
si:ch211-212k5.4
|
si:ch211-212k5.4 |
| chr3_-_22191132 | 0.61 |
ENSDART00000154226
ENSDART00000155528 ENSDART00000155190 |
maptb
|
microtubule-associated protein tau b |
| chr2_+_21452822 | 0.60 |
ENSDART00000169028
|
afg3l2
|
AFG3-like AAA ATPase 2 |
| chr21_+_20771082 | 0.60 |
ENSDART00000079732
|
oxct1b
|
3-oxoacid CoA transferase 1b |
| chr16_+_19013018 | 0.59 |
ENSDART00000191526
|
si:ch211-254p10.2
|
si:ch211-254p10.2 |
| chr20_+_51479263 | 0.59 |
ENSDART00000148798
|
tlr5a
|
toll-like receptor 5a |
| chr23_-_574727 | 0.58 |
ENSDART00000135183
ENSDART00000114827 |
kcnk15
|
potassium channel, subfamily K, member 15 |
| chr13_+_13930263 | 0.58 |
ENSDART00000079154
|
rpia
|
ribose 5-phosphate isomerase A (ribose 5-phosphate epimerase) |
| chr1_+_39553040 | 0.58 |
ENSDART00000137676
|
tenm3
|
teneurin transmembrane protein 3 |
| chr9_+_26199674 | 0.58 |
ENSDART00000139510
|
si:dkey-111i23.1
|
si:dkey-111i23.1 |
| chr7_-_16034324 | 0.57 |
ENSDART00000002498
ENSDART00000162962 |
elp4
|
elongator acetyltransferase complex subunit 4 |
| chr20_-_116999 | 0.57 |
ENSDART00000149581
|
ypel5
|
yippee-like 5 |
| chr17_+_20607487 | 0.56 |
ENSDART00000154123
|
si:ch73-288o11.5
|
si:ch73-288o11.5 |
| chr23_+_42336084 | 0.56 |
ENSDART00000158959
ENSDART00000161812 |
cyp2aa7
cyp2aa8
|
cytochrome P450, family 2, subfamily AA, polypeptide 7 cytochrome P450, family 2, subfamily AA, polypeptide 8 |
| chr7_+_20524064 | 0.56 |
ENSDART00000052917
|
slc3a2a
|
solute carrier family 3 (amino acid transporter heavy chain), member 2a |
| chr8_-_30791266 | 0.56 |
ENSDART00000062220
|
gstt1a
|
glutathione S-transferase theta 1a |
| chr9_+_38427572 | 0.56 |
ENSDART00000108860
|
CYP27A1 (1 of many)
|
zgc:136333 |
| chr7_-_5316901 | 0.56 |
ENSDART00000181505
ENSDART00000124367 |
si:cabz01074946.1
|
si:cabz01074946.1 |
| chr23_-_26120281 | 0.54 |
ENSDART00000133372
ENSDART00000103872 |
rca2.1
|
regulator of complement activation group 2 gene 1 |
| chr1_+_34295925 | 0.54 |
ENSDART00000075584
|
kctd12.2
|
potassium channel tetramerisation domain containing 12.2 |
| chr18_-_14836862 | 0.54 |
ENSDART00000124843
|
mtss1la
|
metastasis suppressor 1-like a |
| chr22_-_36856405 | 0.53 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr19_-_10730488 | 0.53 |
ENSDART00000126033
|
slc9a3.1
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3, tandem duplicate 1 |
| chr15_-_23908779 | 0.52 |
ENSDART00000088808
|
usp32
|
ubiquitin specific peptidase 32 |
| chr8_-_23702295 | 0.52 |
ENSDART00000162296
|
cfp
|
complement factor properdin |
| chr16_+_33902006 | 0.51 |
ENSDART00000161807
ENSDART00000159474 |
gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr3_-_12970418 | 0.51 |
ENSDART00000158747
|
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr8_-_30791089 | 0.51 |
ENSDART00000147332
|
gstt1a
|
glutathione S-transferase theta 1a |
| chr5_-_43935460 | 0.51 |
ENSDART00000166152
ENSDART00000188969 |
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr22_+_26263290 | 0.50 |
ENSDART00000184840
|
BX649315.1
|
|
| chr13_-_18637244 | 0.50 |
ENSDART00000057869
|
mat1a
|
methionine adenosyltransferase I, alpha |
| chr9_+_53327332 | 0.50 |
ENSDART00000125715
|
dct
|
dopachrome tautomerase |
| chr22_-_37611681 | 0.50 |
ENSDART00000028085
|
ttc14
|
tetratricopeptide repeat domain 14 |
| chr8_-_7440364 | 0.50 |
ENSDART00000180659
|
hdac6
|
histone deacetylase 6 |
| chr16_+_5901835 | 0.49 |
ENSDART00000060519
|
ulk4
|
unc-51 like kinase 4 |
| chr14_-_33351103 | 0.49 |
ENSDART00000010019
|
upf3b
|
UPF3B, regulator of nonsense mediated mRNA decay |
| chr3_-_18410968 | 0.49 |
ENSDART00000041842
|
ndufb10
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10 |
| chr21_-_45891262 | 0.49 |
ENSDART00000169816
|
galnt10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
| chr9_+_38399912 | 0.48 |
ENSDART00000022246
ENSDART00000145892 |
bcs1l
|
BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
| chr2_-_58085444 | 0.48 |
ENSDART00000166395
|
fcer1gl
|
Fc receptor, IgE, high affinity I, gamma polypeptide like |
| chr1_+_8593560 | 0.48 |
ENSDART00000161791
|
si:ch211-160d14.15
|
si:ch211-160d14.15 |
| chr17_-_45733401 | 0.48 |
ENSDART00000185727
|
arf6b
|
ADP-ribosylation factor 6b |
| chr11_-_23458792 | 0.47 |
ENSDART00000032844
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr21_+_5882300 | 0.47 |
ENSDART00000165065
|
uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr16_+_37470717 | 0.47 |
ENSDART00000112003
ENSDART00000188431 ENSDART00000192837 |
adgrb1a
|
adhesion G protein-coupled receptor B1a |
| chr4_+_44577872 | 0.46 |
ENSDART00000157214
|
si:dkey-7j22.2
|
si:dkey-7j22.2 |
| chr19_+_27479563 | 0.46 |
ENSDART00000049368
ENSDART00000185426 |
atat1
|
alpha tubulin acetyltransferase 1 |
| chr10_+_42358426 | 0.46 |
ENSDART00000025691
|
dbnla
|
drebrin-like a |
| chr13_+_22480496 | 0.46 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr3_-_54544612 | 0.45 |
ENSDART00000018044
|
angptl6
|
angiopoietin-like 6 |
| chr25_+_34014523 | 0.45 |
ENSDART00000182856
|
anxa2a
|
annexin A2a |
| chr4_-_32180155 | 0.44 |
ENSDART00000164151
|
si:dkey-72l17.6
|
si:dkey-72l17.6 |
| chr4_+_67050047 | 0.44 |
ENSDART00000170257
|
znf1070
|
zinc finger protein 1070 |
| chr9_-_44295071 | 0.43 |
ENSDART00000011837
|
neurod1
|
neuronal differentiation 1 |
| chr14_+_41576849 | 0.43 |
ENSDART00000074424
|
nitr13
|
novel immune-type receptor 13 |
| chr10_-_40589641 | 0.42 |
ENSDART00000140228
ENSDART00000154850 |
taar16b
|
trace amine associated receptor 16b |
| chr8_+_24296639 | 0.42 |
ENSDART00000172002
|
ZNF335
|
si:ch211-269m15.3 |
| chr4_+_58057168 | 0.42 |
ENSDART00000161097
|
si:dkey-57k17.1
|
si:dkey-57k17.1 |
| chr2_+_3809226 | 0.42 |
ENSDART00000147261
|
egfra
|
epidermal growth factor receptor a (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) |
| chr20_-_48470599 | 0.42 |
ENSDART00000166857
|
CABZ01059120.1
|
|
| chr6_-_28345002 | 0.42 |
ENSDART00000158955
|
si:busm1-105l16.2
|
si:busm1-105l16.2 |
| chr7_-_6427198 | 0.41 |
ENSDART00000173446
|
FP340257.1
|
|
| chr8_+_44613135 | 0.41 |
ENSDART00000063392
|
lsm1
|
LSM1, U6 small nuclear RNA associated |
| chr20_+_34870517 | 0.41 |
ENSDART00000191797
|
ankef1a
|
ankyrin repeat and EF-hand domain containing 1a |
| chr19_+_19600297 | 0.40 |
ENSDART00000160134
ENSDART00000183493 |
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr19_+_27479838 | 0.40 |
ENSDART00000103922
|
atat1
|
alpha tubulin acetyltransferase 1 |
| chr2_-_11912347 | 0.39 |
ENSDART00000023851
|
abhd3
|
abhydrolase domain containing 3 |
| chr4_-_36522557 | 0.39 |
ENSDART00000167619
|
BX649453.1
|
Danio rerio gastrula zinc finger protein XlCGF8.2DB-like (LOC100150619), mRNA. |
| chr21_-_21089781 | 0.39 |
ENSDART00000144361
|
ank1b
|
ankyrin 1, erythrocytic b |
| chr2_+_29976419 | 0.38 |
ENSDART00000056748
|
en2b
|
engrailed homeobox 2b |
| chr4_-_36144500 | 0.38 |
ENSDART00000170896
|
znf992
|
zinc finger protein 992 |
| chr14_+_34966598 | 0.37 |
ENSDART00000004550
|
rnf145a
|
ring finger protein 145a |
| chr18_+_29402623 | 0.37 |
ENSDART00000014703
|
mafa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog a (paralog a) |
| chr3_-_39198113 | 0.37 |
ENSDART00000102690
|
RETSAT
|
zgc:154169 |
| chr22_+_34663328 | 0.37 |
ENSDART00000183912
|
si:ch73-304f21.1
|
si:ch73-304f21.1 |
| chr11_+_25681335 | 0.37 |
ENSDART00000121478
ENSDART00000190877 |
otud5b
|
OTU deubiquitinase 5b |
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb47b+znf652
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.4 | GO:1904355 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.8 | 2.4 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.5 | 2.4 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.3 | 1.0 | GO:1903961 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.3 | 1.0 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 0.3 | 1.3 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.3 | 2.0 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.2 | 1.2 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.2 | 0.7 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) vitamin D biosynthetic process(GO:0042368) cellular alcohol metabolic process(GO:0044107) cellular alcohol biosynthetic process(GO:0044108) |
| 0.2 | 1.7 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.2 | 0.6 | GO:1905133 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.2 | 1.4 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 0.8 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.2 | 1.6 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.2 | 1.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 1.3 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.2 | 0.5 | GO:0055057 | neuroblast division(GO:0055057) positive regulation of stem cell proliferation(GO:2000648) |
| 0.2 | 1.6 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.1 | 0.6 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.6 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.1 | 0.5 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 0.6 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.4 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 1.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.6 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 | 1.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.7 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 1.7 | GO:0042119 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.1 | 0.3 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.5 | GO:0051204 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 1.0 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.5 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 1.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.2 | GO:0071706 | tumor necrosis factor production(GO:0032640) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 0.1 | 0.4 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.1 | 1.3 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.3 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 0.5 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 0.7 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.1 | 0.6 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 0.9 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 0.5 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.8 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 0.4 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 1.3 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.3 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.1 | 3.9 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.1 | 0.9 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.4 | GO:0006699 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.1 | 0.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.4 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.5 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.6 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.1 | 1.7 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.1 | 0.3 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.2 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 1.0 | GO:0006949 | syncytium formation by plasma membrane fusion(GO:0000768) syncytium formation(GO:0006949) |
| 0.1 | 1.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.5 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 1.3 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 1.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.5 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.0 | 0.6 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.2 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.7 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.5 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.2 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.0 | 0.8 | GO:0019229 | regulation of vasoconstriction(GO:0019229) |
| 0.0 | 0.6 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.1 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.0 | 0.3 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.4 | GO:0072554 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 0.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.3 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.4 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.0 | 0.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 1.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.5 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 7.0 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.5 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.5 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.9 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.1 | GO:0045190 | B cell activation involved in immune response(GO:0002312) isotype switching(GO:0045190) |
| 0.0 | 0.1 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.3 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 0.2 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 1.7 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.5 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.3 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 2.5 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 1.1 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.0 | 0.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 1.3 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.1 | GO:0051601 | exocyst localization(GO:0051601) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.9 | GO:0016586 | RSC complex(GO:0016586) |
| 0.2 | 0.9 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.2 | 1.3 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.2 | 2.4 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 0.5 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.1 | 1.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 1.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.9 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.9 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.9 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 1.5 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 0.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.4 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.5 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 1.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.7 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.5 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.4 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.4 | 1.7 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.3 | 1.3 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.3 | 0.9 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.2 | 0.7 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) cholesterol 26-hydroxylase activity(GO:0031073) |
| 0.2 | 0.9 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 0.6 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.2 | 0.8 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.2 | 1.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 2.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.2 | 1.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 1.3 | GO:0030291 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.2 | 0.5 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) IgE receptor activity(GO:0019767) |
| 0.2 | 0.8 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.4 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 0.5 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 0.5 | GO:0047611 | tubulin deacetylase activity(GO:0042903) acetylspermidine deacetylase activity(GO:0047611) |
| 0.1 | 0.6 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 0.3 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.1 | 0.4 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 1.0 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.3 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.1 | 1.5 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 1.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.5 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.3 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.1 | 3.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.6 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.6 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 0.8 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 1.8 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 1.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 1.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 1.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.5 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.2 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 1.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.0 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.4 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 1.1 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.7 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.8 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 1.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.6 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.4 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.9 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 0.4 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.2 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.7 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.8 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 0.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 0.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 1.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 2.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.4 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |