Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
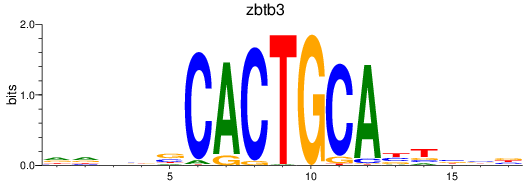
Results for zbtb3
Z-value: 0.53
Transcription factors associated with zbtb3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zbtb3
|
ENSDARG00000036235 | zinc finger and BTB domain containing 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zbtb3 | dr11_v1_chr14_+_35405518_35405518 | -0.47 | 5.1e-02 | Click! |
Activity profile of zbtb3 motif
Sorted Z-values of zbtb3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_52398205 | 2.96 |
ENSDART00000143225
|
si:ch211-217k17.9
|
si:ch211-217k17.9 |
| chr6_+_13039951 | 1.33 |
ENSDART00000091700
|
catip
|
ciliogenesis associated TTC17 interacting protein |
| chr23_-_32404022 | 1.28 |
ENSDART00000156387
ENSDART00000155508 |
si:ch211-66i15.4
|
si:ch211-66i15.4 |
| chr22_-_25469751 | 1.15 |
ENSDART00000171670
|
CR769772.4
|
|
| chr14_+_6546516 | 1.02 |
ENSDART00000097214
|
adam19b
|
ADAM metallopeptidase domain 19b |
| chr1_+_50921266 | 0.98 |
ENSDART00000006538
|
otx2a
|
orthodenticle homeobox 2a |
| chr25_+_13662606 | 0.87 |
ENSDART00000008989
|
ccdc113
|
coiled-coil domain containing 113 |
| chr22_-_25502977 | 0.86 |
ENSDART00000181749
|
CR769772.4
|
|
| chr1_-_44704261 | 0.78 |
ENSDART00000133210
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr18_+_7611298 | 0.73 |
ENSDART00000062156
|
odf3b
|
outer dense fiber of sperm tails 3B |
| chr14_-_1355544 | 0.70 |
ENSDART00000060417
|
cetn4
|
centrin 4 |
| chr16_-_32013913 | 0.63 |
ENSDART00000030282
ENSDART00000138701 |
gstk1
|
glutathione S-transferase kappa 1 |
| chr16_+_32092866 | 0.56 |
ENSDART00000137785
|
si:dkey-40m6.11
|
si:dkey-40m6.11 |
| chr5_+_9297585 | 0.46 |
ENSDART00000170557
|
si:ch73-194c23.1
|
si:ch73-194c23.1 |
| chr13_-_2215213 | 0.43 |
ENSDART00000129773
|
mlip
|
muscular LMNA-interacting protein |
| chr3_+_24458899 | 0.42 |
ENSDART00000156655
|
cbx6b
|
chromobox homolog 6b |
| chr3_-_24458281 | 0.42 |
ENSDART00000153993
|
baiap2l2a
|
BAI1-associated protein 2-like 2a |
| chr22_-_18387059 | 0.40 |
ENSDART00000007769
|
tssk6
|
testis-specific serine kinase 6 |
| chr7_-_35432901 | 0.38 |
ENSDART00000026712
|
mmp2
|
matrix metallopeptidase 2 |
| chr19_-_7240627 | 0.37 |
ENSDART00000181874
|
FP101875.2
|
|
| chr12_-_35885349 | 0.36 |
ENSDART00000085662
|
cep131
|
centrosomal protein 131 |
| chr20_+_25340814 | 0.36 |
ENSDART00000063028
|
ctgfa
|
connective tissue growth factor a |
| chr13_-_37127970 | 0.35 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr2_-_985417 | 0.34 |
ENSDART00000140540
|
si:ch211-241e1.3
|
si:ch211-241e1.3 |
| chr3_+_24458204 | 0.30 |
ENSDART00000155028
ENSDART00000153551 |
cbx6b
|
chromobox homolog 6b |
| chr17_-_11151655 | 0.30 |
ENSDART00000156383
|
CU179699.1
|
|
| chr10_+_44641599 | 0.29 |
ENSDART00000172128
|
sez6l
|
seizure related 6 homolog (mouse)-like |
| chr9_-_8979154 | 0.28 |
ENSDART00000145266
|
ing1
|
inhibitor of growth family, member 1 |
| chr19_-_19442983 | 0.28 |
ENSDART00000052649
|
sb:cb649
|
sb:cb649 |
| chr21_+_45366229 | 0.28 |
ENSDART00000029946
|
ube2b
|
ubiquitin-conjugating enzyme E2B (RAD6 homolog) |
| chr14_-_32258759 | 0.27 |
ENSDART00000052949
|
fgf13a
|
fibroblast growth factor 13a |
| chr7_-_25895189 | 0.27 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr7_+_34492744 | 0.26 |
ENSDART00000109635
ENSDART00000173844 |
calml4a
|
calmodulin-like 4a |
| chr20_+_39375280 | 0.26 |
ENSDART00000172434
|
pimr131
|
Pim proto-oncogene, serine/threonine kinase, related 131 |
| chr14_-_23801389 | 0.25 |
ENSDART00000054264
|
nr3c1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr1_-_54425791 | 0.25 |
ENSDART00000039911
|
pkd1a
|
polycystic kidney disease 1a |
| chr10_-_4375190 | 0.24 |
ENSDART00000016102
|
CABZ01073795.1
|
|
| chr7_-_26087807 | 0.24 |
ENSDART00000052989
|
ache
|
acetylcholinesterase |
| chr16_+_4695075 | 0.23 |
ENSDART00000039054
|
mecr
|
mitochondrial trans-2-enoyl-CoA reductase |
| chr21_-_4923427 | 0.22 |
ENSDART00000181489
|
kcnt1
|
potassium channel, subfamily T, member 1 |
| chr16_-_42872571 | 0.21 |
ENSDART00000154757
ENSDART00000102345 |
txnipb
|
thioredoxin interacting protein b |
| chr13_+_771403 | 0.21 |
ENSDART00000093166
|
nrxn1b
|
neurexin 1b |
| chr20_+_38671894 | 0.20 |
ENSDART00000146544
|
mpv17
|
MpV17 mitochondrial inner membrane protein |
| chr18_+_37015185 | 0.19 |
ENSDART00000191305
|
sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chr23_-_29878643 | 0.19 |
ENSDART00000058407
|
slc25a33
|
solute carrier family 25 (pyrimidine nucleotide carrier), member 33 |
| chr3_+_53240562 | 0.18 |
ENSDART00000031234
|
stxbp2
|
syntaxin binding protein 2 |
| chr19_+_917852 | 0.17 |
ENSDART00000082466
|
tgfbr2a
|
transforming growth factor beta receptor 2a |
| chr4_-_68913650 | 0.16 |
ENSDART00000184297
|
si:dkey-264f17.5
|
si:dkey-264f17.5 |
| chr1_-_633356 | 0.16 |
ENSDART00000171019
|
appa
|
amyloid beta (A4) precursor protein a |
| chr16_+_10422836 | 0.15 |
ENSDART00000161568
|
ino80e
|
INO80 complex subunit E |
| chr24_-_26383978 | 0.15 |
ENSDART00000031426
|
skilb
|
SKI-like proto-oncogene b |
| chr5_+_61799629 | 0.14 |
ENSDART00000113508
|
hnrnpul1l
|
heterogeneous nuclear ribonucleoprotein U-like 1 like |
| chr19_-_35439237 | 0.14 |
ENSDART00000145883
|
anln
|
anillin, actin binding protein |
| chr12_+_48220584 | 0.14 |
ENSDART00000164392
|
lrrc20
|
leucine rich repeat containing 20 |
| chr10_+_26944978 | 0.14 |
ENSDART00000192148
|
frmd8
|
FERM domain containing 8 |
| chr20_-_6131686 | 0.14 |
ENSDART00000145964
ENSDART00000086578 ENSDART00000164090 |
pum2
|
pumilio RNA-binding family member 2 |
| chr3_-_34069637 | 0.14 |
ENSDART00000151588
|
ighv9-1
|
immunoglobulin heavy variable 9-1 |
| chr8_-_54304381 | 0.12 |
ENSDART00000184177
|
RHO (1 of many)
|
rhodopsin |
| chr4_+_38550788 | 0.12 |
ENSDART00000157412
|
si:ch211-209n20.1
|
si:ch211-209n20.1 |
| chr14_+_1355857 | 0.11 |
ENSDART00000188008
|
bbs12
|
Bardet-Biedl syndrome 12 |
| chr2_-_27329214 | 0.11 |
ENSDART00000145835
|
tmx3a
|
thioredoxin related transmembrane protein 3a |
| chr21_+_35296246 | 0.10 |
ENSDART00000076750
|
il12bb
|
interleukin 12B, b |
| chr10_+_31951338 | 0.10 |
ENSDART00000019416
|
lhfpl6
|
LHFPL tetraspan subfamily member 6 |
| chr8_+_3820134 | 0.10 |
ENSDART00000122454
|
citb
|
citron rho-interacting serine/threonine kinase b |
| chr3_-_61185746 | 0.10 |
ENSDART00000028219
|
pvalb4
|
parvalbumin 4 |
| chr8_-_1884955 | 0.10 |
ENSDART00000081563
|
si:dkey-178e17.3
|
si:dkey-178e17.3 |
| chr18_-_43880020 | 0.09 |
ENSDART00000185638
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr18_-_50839033 | 0.08 |
ENSDART00000169773
|
ddb1
|
damage-specific DNA binding protein 1 |
| chr23_+_39558508 | 0.08 |
ENSDART00000017902
|
camk1gb
|
calcium/calmodulin-dependent protein kinase IGb |
| chr3_+_61185660 | 0.08 |
ENSDART00000167114
|
CR352263.1
|
|
| chr22_-_29336268 | 0.08 |
ENSDART00000132776
ENSDART00000186351 ENSDART00000121599 |
pdgfba
|
platelet-derived growth factor beta polypeptide a |
| chr17_+_34215886 | 0.08 |
ENSDART00000186775
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr2_+_5621529 | 0.07 |
ENSDART00000144187
|
fgf12a
|
fibroblast growth factor 12a |
| chr22_+_3238474 | 0.07 |
ENSDART00000157954
|
si:ch1073-178p5.3
|
si:ch1073-178p5.3 |
| chr19_+_42770041 | 0.07 |
ENSDART00000150930
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr8_+_26293673 | 0.06 |
ENSDART00000144977
|
mgll
|
monoglyceride lipase |
| chr6_-_51556308 | 0.06 |
ENSDART00000149033
|
rbl1
|
retinoblastoma-like 1 (p107) |
| chr17_-_29311835 | 0.06 |
ENSDART00000104224
|
tecpr2
|
tectonin beta-propeller repeat containing 2 |
| chr1_+_532766 | 0.05 |
ENSDART00000179731
ENSDART00000060944 |
mrpl39
|
mitochondrial ribosomal protein L39 |
| chr2_+_45643122 | 0.04 |
ENSDART00000138179
|
fndc7rs4
|
fibronectin type III domain containing 7, related sequence 4 |
| chr15_-_576135 | 0.04 |
ENSDART00000124170
|
cbln20
|
cerebellin 20 |
| chr18_+_7309150 | 0.04 |
ENSDART00000052792
|
cers3b
|
ceramide synthase 3b |
| chr11_+_1805421 | 0.04 |
ENSDART00000173143
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr2_+_45484183 | 0.04 |
ENSDART00000183490
|
si:ch211-66k16.28
|
si:ch211-66k16.28 |
| chr22_-_8353386 | 0.03 |
ENSDART00000077600
|
CABZ01061495.1
|
|
| chr24_-_24146875 | 0.03 |
ENSDART00000173052
|
map7d2b
|
MAP7 domain containing 2b |
| chr25_-_35542739 | 0.03 |
ENSDART00000097651
|
si:ch211-87j1.4
|
si:ch211-87j1.4 |
| chr11_-_36156935 | 0.03 |
ENSDART00000124935
ENSDART00000138609 |
brk1
gpx1a
|
BRICK1, SCAR/WAVE actin-nucleating complex subunit glutathione peroxidase 1a |
| chr16_+_34531486 | 0.03 |
ENSDART00000043291
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr9_-_32753535 | 0.03 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr15_+_863931 | 0.03 |
ENSDART00000154083
|
si:dkey-7i4.12
|
si:dkey-7i4.12 |
| chr1_-_22687913 | 0.03 |
ENSDART00000168171
|
fgfbp2b
|
fibroblast growth factor binding protein 2b |
| chr6_-_40921412 | 0.03 |
ENSDART00000076097
ENSDART00000188364 |
sfi1
|
SFI1 centrin binding protein |
| chr5_+_53580846 | 0.03 |
ENSDART00000184967
ENSDART00000161751 |
taf6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr13_+_23282549 | 0.03 |
ENSDART00000101134
|
khdrbs2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr6_-_38419318 | 0.02 |
ENSDART00000138026
|
gabra5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr22_-_11136625 | 0.02 |
ENSDART00000016873
ENSDART00000125561 |
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr7_-_6592142 | 0.02 |
ENSDART00000160137
|
kcnj10a
|
potassium inwardly-rectifying channel, subfamily J, member 10a |
| chr6_-_39006449 | 0.02 |
ENSDART00000150885
|
vdrb
|
vitamin D receptor b |
| chr5_-_20446082 | 0.02 |
ENSDART00000051607
|
ISCU (1 of many)
|
si:ch211-191d15.2 |
| chr16_-_14552199 | 0.02 |
ENSDART00000133368
|
si:dkey-237j11.3
|
si:dkey-237j11.3 |
| chr1_+_59007536 | 0.01 |
ENSDART00000165339
|
CDC37
|
cell division cycle 37 |
| chr7_+_17938128 | 0.01 |
ENSDART00000141044
|
mta2
|
metastasis associated 1 family, member 2 |
| chr15_-_19705707 | 0.00 |
ENSDART00000047643
|
sytl2b
|
synaptotagmin-like 2b |
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.1 | 0.2 | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.1 | 0.3 | GO:0071548 | cellular response to cortisol stimulus(GO:0071387) response to dexamethasone(GO:0071548) |
| 0.1 | 0.3 | GO:0032965 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) |
| 0.1 | 0.2 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.2 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.1 | 1.0 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.4 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 1.0 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.4 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.7 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0043514 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 1.3 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.7 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.4 | GO:0016605 | PML body(GO:0016605) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.2 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0042164 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.0 | 1.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |