Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for zbtb16a+zbtb16b
Z-value: 0.21
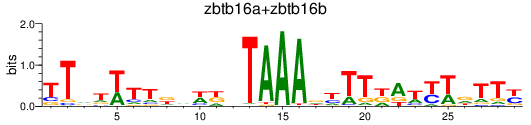
Transcription factors associated with zbtb16a+zbtb16b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zbtb16a
|
ENSDARG00000007184 | zinc finger and BTB domain containing 16a |
|
zbtb16b
|
ENSDARG00000074526 | zinc finger and BTB domain containing 16b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zbtb16a | dr11_v1_chr21_-_23308286_23308286 | 0.62 | 5.9e-03 | Click! |
| zbtb16b | dr11_v1_chr15_-_18361475_18361475 | -0.00 | 9.8e-01 | Click! |
Activity profile of zbtb16a+zbtb16b motif
Sorted Z-values of zbtb16a+zbtb16b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_72003301 | 0.34 |
ENSDART00000012918
ENSDART00000182268 ENSDART00000185750 |
psmd9
|
proteasome 26S subunit, non-ATPase 9 |
| chr12_-_45197038 | 0.33 |
ENSDART00000016635
|
bccip
|
BRCA2 and CDKN1A interacting protein |
| chr9_+_52398531 | 0.32 |
ENSDART00000126215
|
dap1b
|
death associated protein 1b |
| chr12_+_2870671 | 0.25 |
ENSDART00000165225
|
prkar1ab
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) b |
| chr18_-_34170918 | 0.22 |
ENSDART00000015079
|
slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr18_-_34171280 | 0.20 |
ENSDART00000122321
|
slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr5_-_68826177 | 0.19 |
ENSDART00000136605
|
si:ch211-283h6.4
|
si:ch211-283h6.4 |
| chr6_+_3730843 | 0.19 |
ENSDART00000019630
|
FO704755.1
|
|
| chr5_+_2693601 | 0.18 |
ENSDART00000183882
|
rabepk
|
Rab9 effector protein with kelch motifs |
| chr19_+_636886 | 0.18 |
ENSDART00000149192
|
tert
|
telomerase reverse transcriptase |
| chr21_-_43636595 | 0.17 |
ENSDART00000151115
ENSDART00000151486 ENSDART00000151778 |
si:ch1073-263o8.2
|
si:ch1073-263o8.2 |
| chr6_-_49510553 | 0.16 |
ENSDART00000166238
|
rplp2
|
ribosomal protein, large P2 |
| chr20_-_16171297 | 0.16 |
ENSDART00000012476
|
coa7
|
cytochrome c oxidase assembly factor 7 |
| chr21_+_22846757 | 0.15 |
ENSDART00000185766
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr5_-_67292690 | 0.15 |
ENSDART00000062366
|
rsrc2
|
arginine/serine-rich coiled-coil 2 |
| chr20_+_26880668 | 0.15 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr23_+_19198244 | 0.14 |
ENSDART00000047015
|
ccdc115
|
coiled-coil domain containing 115 |
| chr21_-_13225402 | 0.14 |
ENSDART00000080347
|
wdr34
|
WD repeat domain 34 |
| chr2_-_122154 | 0.14 |
ENSDART00000156248
ENSDART00000004071 |
znfl2a
|
zinc finger-like gene 2a |
| chr18_+_30508729 | 0.13 |
ENSDART00000185140
|
cox4i1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr2_-_23931536 | 0.13 |
ENSDART00000121885
|
tgfbr1a
|
transforming growth factor, beta receptor 1 a |
| chr3_+_7822948 | 0.13 |
ENSDART00000169921
|
trir
|
telomerase RNA component interacting RNase |
| chr21_+_10712823 | 0.13 |
ENSDART00000123476
|
lman1
|
lectin, mannose-binding, 1 |
| chr14_-_14659023 | 0.13 |
ENSDART00000170355
ENSDART00000159888 ENSDART00000172241 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr2_+_16160906 | 0.13 |
ENSDART00000135783
|
selenoj
|
selenoprotein J |
| chr7_-_24520866 | 0.13 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr25_+_21716263 | 0.12 |
ENSDART00000148920
|
znf277
|
zinc finger protein 277 |
| chr24_+_40860320 | 0.12 |
ENSDART00000161351
|
gorasp1b
|
golgi reassembly stacking protein 1b |
| chr15_-_25584888 | 0.12 |
ENSDART00000127571
|
si:dkey-54n8.2
|
si:dkey-54n8.2 |
| chr2_-_7757273 | 0.12 |
ENSDART00000136074
|
b3gnt5b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5b |
| chr12_+_3871452 | 0.12 |
ENSDART00000066546
|
nif3l1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr6_-_30689126 | 0.11 |
ENSDART00000065211
|
pars2
|
prolyl-tRNA synthetase 2, mitochondrial |
| chr18_+_38755023 | 0.11 |
ENSDART00000010177
ENSDART00000193136 |
onecut1
|
one cut homeobox 1 |
| chr23_-_31506854 | 0.11 |
ENSDART00000131352
ENSDART00000138625 ENSDART00000133002 |
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr10_+_7636811 | 0.11 |
ENSDART00000160673
|
hint1
|
histidine triad nucleotide binding protein 1 |
| chr24_-_36175365 | 0.10 |
ENSDART00000065338
|
pak1ip1
|
PAK1 interacting protein 1 |
| chr13_+_20007366 | 0.10 |
ENSDART00000147217
|
atrnl1a
|
attractin-like 1a |
| chr2_+_6126638 | 0.10 |
ENSDART00000153916
|
fzr1b
|
fizzy/cell division cycle 20 related 1b |
| chr1_+_49652967 | 0.10 |
ENSDART00000191296
|
tsga10
|
testis specific, 10 |
| chr5_-_31716713 | 0.10 |
ENSDART00000131443
|
dpm2
|
dolichyl-phosphate mannosyltransferase polypeptide 2, regulatory subunit |
| chr11_-_3954691 | 0.10 |
ENSDART00000182041
|
pbrm1
|
polybromo 1 |
| chr24_-_41478917 | 0.09 |
ENSDART00000192192
|
CABZ01084131.1
|
|
| chr4_+_5796761 | 0.09 |
ENSDART00000164854
|
si:ch73-352p4.8
|
si:ch73-352p4.8 |
| chr16_-_35579200 | 0.09 |
ENSDART00000162172
|
scmh1
|
Scm polycomb group protein homolog 1 |
| chr11_-_12364122 | 0.09 |
ENSDART00000170129
|
zgc:174353
|
zgc:174353 |
| chr1_+_38362412 | 0.09 |
ENSDART00000075086
|
cep44
|
centrosomal protein 44 |
| chr2_+_6126086 | 0.09 |
ENSDART00000179962
|
fzr1b
|
fizzy/cell division cycle 20 related 1b |
| chr2_-_15033136 | 0.09 |
ENSDART00000145974
ENSDART00000034865 |
hccsa.1
|
holocytochrome c synthase a |
| chr19_-_27588842 | 0.09 |
ENSDART00000121643
|
si:dkeyp-46h3.2
|
si:dkeyp-46h3.2 |
| chr7_+_9873750 | 0.08 |
ENSDART00000173348
ENSDART00000172813 |
lins1
|
lines homolog 1 |
| chr19_-_3876877 | 0.08 |
ENSDART00000163711
|
thrap3b
|
thyroid hormone receptor associated protein 3b |
| chr23_-_38160024 | 0.08 |
ENSDART00000087112
|
pfdn4
|
prefoldin subunit 4 |
| chr20_+_36628059 | 0.08 |
ENSDART00000062898
|
ephx1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr24_-_24118451 | 0.08 |
ENSDART00000111096
|
zgc:112982
|
zgc:112982 |
| chr10_+_24690534 | 0.08 |
ENSDART00000079549
|
tpte
|
transmembrane phosphatase with tensin homology |
| chr22_+_10159512 | 0.08 |
ENSDART00000184415
|
rpp14
|
ribonuclease P/MRP 14 subunit |
| chr11_-_12530304 | 0.08 |
ENSDART00000143061
|
zgc:174354
|
zgc:174354 |
| chr4_+_2230701 | 0.08 |
ENSDART00000080439
|
cmah
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| chr20_+_16721933 | 0.08 |
ENSDART00000063950
|
psmc1b
|
proteasome 26S subunit, ATPase 1b |
| chr7_-_28568238 | 0.08 |
ENSDART00000173927
|
tmem9b
|
TMEM9 domain family, member B |
| chr14_-_21123551 | 0.08 |
ENSDART00000171679
ENSDART00000165882 |
si:dkey-74k8.4
|
si:dkey-74k8.4 |
| chr3_+_32125452 | 0.08 |
ENSDART00000110396
|
zgc:194125
|
zgc:194125 |
| chr12_-_20796430 | 0.07 |
ENSDART00000064339
|
nme2a
|
NME/NM23 nucleoside diphosphate kinase 2a |
| chr21_-_293146 | 0.07 |
ENSDART00000157781
|
plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr25_+_19541019 | 0.07 |
ENSDART00000141056
ENSDART00000027740 |
nedd1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr16_-_54800202 | 0.07 |
ENSDART00000059560
ENSDART00000161833 |
khdc4
|
KH domain containing 4, pre-mRNA splicing factor |
| chr22_+_1421212 | 0.07 |
ENSDART00000161813
|
zgc:101130
|
zgc:101130 |
| chr22_-_9889413 | 0.07 |
ENSDART00000081450
|
si:dkey-253d23.5
|
si:dkey-253d23.5 |
| chr23_+_42810055 | 0.07 |
ENSDART00000186647
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr8_+_24296639 | 0.07 |
ENSDART00000172002
|
ZNF335
|
si:ch211-269m15.3 |
| chr11_-_12379541 | 0.07 |
ENSDART00000171717
|
zgc:174353
|
zgc:174353 |
| chr10_+_14499201 | 0.07 |
ENSDART00000064901
|
katnal2
|
katanin p60 subunit A-like 2 |
| chr22_-_27115241 | 0.07 |
ENSDART00000019442
|
xpnpep1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr16_+_36126310 | 0.07 |
ENSDART00000166040
ENSDART00000189802 |
sh3bp5b
|
SH3-domain binding protein 5b (BTK-associated) |
| chr8_+_26083808 | 0.07 |
ENSDART00000099283
|
dalrd3
|
DALR anticodon binding domain containing 3 |
| chr22_-_5252005 | 0.07 |
ENSDART00000132942
ENSDART00000081801 |
ncln
|
nicalin |
| chr12_+_10448226 | 0.06 |
ENSDART00000152824
|
crym
|
crystallin, mu |
| chr2_-_1548330 | 0.06 |
ENSDART00000082155
ENSDART00000108481 ENSDART00000111272 |
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr17_-_18888959 | 0.06 |
ENSDART00000080029
|
ak7b
|
adenylate kinase 7b |
| chr9_+_8894788 | 0.06 |
ENSDART00000132068
|
naxd
|
NAD(P)HX dehydratase |
| chr17_+_21295132 | 0.06 |
ENSDART00000103845
|
eno4
|
enolase family member 4 |
| chr6_-_51556308 | 0.06 |
ENSDART00000149033
|
rbl1
|
retinoblastoma-like 1 (p107) |
| chr25_+_34888886 | 0.06 |
ENSDART00000035245
|
spire2
|
spire-type actin nucleation factor 2 |
| chr21_-_2348838 | 0.06 |
ENSDART00000160337
|
si:ch73-299h12.8
|
si:ch73-299h12.8 |
| chr6_-_21582444 | 0.06 |
ENSDART00000151339
|
si:dkey-43k4.3
|
si:dkey-43k4.3 |
| chr22_-_27113332 | 0.06 |
ENSDART00000146951
ENSDART00000178855 |
xpnpep1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr3_+_60277300 | 0.06 |
ENSDART00000170977
|
sec14l1
|
SEC14-like lipid binding 1 |
| chr5_+_20103085 | 0.06 |
ENSDART00000079424
|
si:rp71-1c23.3
|
si:rp71-1c23.3 |
| chr21_-_25623342 | 0.06 |
ENSDART00000101201
|
tmem223
|
transmembrane protein 223 |
| chr3_-_52683241 | 0.06 |
ENSDART00000155248
|
si:dkey-210j14.5
|
si:dkey-210j14.5 |
| chr11_+_41459408 | 0.06 |
ENSDART00000182285
|
park7
|
parkinson protein 7 |
| chr15_-_17960228 | 0.06 |
ENSDART00000155898
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr17_-_8862424 | 0.06 |
ENSDART00000064633
|
nkl.4
|
NK-lysin tandem duplicate 4 |
| chr12_+_33460794 | 0.05 |
ENSDART00000007053
ENSDART00000142716 |
narf
|
nuclear prelamin A recognition factor |
| chr19_+_27589201 | 0.05 |
ENSDART00000182060
|
si:dkeyp-46h3.1
|
si:dkeyp-46h3.1 |
| chr10_-_7666810 | 0.05 |
ENSDART00000191479
|
pcyox1
|
prenylcysteine oxidase 1 |
| chr22_-_22242884 | 0.05 |
ENSDART00000020937
|
hdgfl2
|
HDGF like 2 |
| chr14_-_24095414 | 0.05 |
ENSDART00000172747
ENSDART00000173146 |
si:ch211-277c7.7
|
si:ch211-277c7.7 |
| chr17_-_31579715 | 0.05 |
ENSDART00000110167
ENSDART00000191092 |
rpap1
|
RNA polymerase II associated protein 1 |
| chr13_-_24332495 | 0.05 |
ENSDART00000140842
|
si:ch211-202m22.1
|
si:ch211-202m22.1 |
| chr11_+_24820542 | 0.05 |
ENSDART00000135443
|
kdm5ba
|
lysine (K)-specific demethylase 5Ba |
| chr6_+_55819038 | 0.05 |
ENSDART00000108786
|
si:ch211-81n22.1
|
si:ch211-81n22.1 |
| chr9_-_34882516 | 0.05 |
ENSDART00000011163
|
asmtl
|
acetylserotonin O-methyltransferase-like |
| chr23_-_4925641 | 0.05 |
ENSDART00000140861
ENSDART00000060718 |
taz
|
tafazzin |
| chr9_+_34232503 | 0.05 |
ENSDART00000132836
|
nxpe3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr12_-_20796658 | 0.05 |
ENSDART00000181253
|
nme2a
|
NME/NM23 nucleoside diphosphate kinase 2a |
| chr4_+_9360532 | 0.05 |
ENSDART00000189971
|
ipo8
|
importin 8 |
| chr9_+_9441453 | 0.05 |
ENSDART00000081859
ENSDART00000188567 ENSDART00000143101 |
maats1
|
MYCBP-associated, testis expressed 1 |
| chr21_-_2341937 | 0.04 |
ENSDART00000158459
|
zgc:193790
|
zgc:193790 |
| chr9_-_18568927 | 0.04 |
ENSDART00000084668
|
enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr25_-_35104083 | 0.04 |
ENSDART00000183252
ENSDART00000156727 |
si:dkey-108k21.21
|
si:dkey-108k21.21 |
| chr12_+_48841182 | 0.04 |
ENSDART00000109315
ENSDART00000185609 ENSDART00000187217 |
dlg5b.1
|
discs, large homolog 5b (Drosophila), tandem duplicate 1 |
| chr20_-_7080427 | 0.04 |
ENSDART00000140166
ENSDART00000023677 |
si:ch211-121a2.2
|
si:ch211-121a2.2 |
| chr25_-_29134654 | 0.04 |
ENSDART00000067066
|
parp6b
|
poly (ADP-ribose) polymerase family, member 6b |
| chr11_-_1509773 | 0.04 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr21_+_45510448 | 0.04 |
ENSDART00000160494
ENSDART00000167914 |
fnip1
|
folliculin interacting protein 1 |
| chr4_+_72612792 | 0.04 |
ENSDART00000106253
|
si:cabz01054394.7
|
si:cabz01054394.7 |
| chr5_-_10768258 | 0.04 |
ENSDART00000157043
|
rtn4r
|
reticulon 4 receptor |
| chr11_+_16040680 | 0.04 |
ENSDART00000157703
|
agtrap
|
angiotensin II receptor-associated protein |
| chr21_-_9383974 | 0.04 |
ENSDART00000160932
|
sdad1
|
SDA1 domain containing 1 |
| chr8_+_31716872 | 0.04 |
ENSDART00000161121
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr24_+_26140855 | 0.04 |
ENSDART00000139017
|
tmtopsb
|
teleost multiple tissue opsin b |
| chr16_+_4838808 | 0.04 |
ENSDART00000179363
|
ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr11_-_889845 | 0.04 |
ENSDART00000162152
|
atg7
|
ATG7 autophagy related 7 homolog (S. cerevisiae) |
| chr16_+_51159775 | 0.04 |
ENSDART00000161924
|
dhdds
|
dehydrodolichyl diphosphate synthase |
| chr16_+_42829735 | 0.04 |
ENSDART00000014956
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr14_+_49376011 | 0.04 |
ENSDART00000020961
|
tnip1
|
TNFAIP3 interacting protein 1 |
| chr22_+_24646339 | 0.04 |
ENSDART00000183865
|
lpar3
|
lysophosphatidic acid receptor 3 |
| chr4_-_17725008 | 0.04 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr12_+_48841419 | 0.04 |
ENSDART00000125331
|
dlg5b.1
|
discs, large homolog 5b (Drosophila), tandem duplicate 1 |
| chr9_+_6592710 | 0.04 |
ENSDART00000179893
|
fhl2a
|
four and a half LIM domains 2a |
| chr1_+_34203817 | 0.04 |
ENSDART00000191432
ENSDART00000046094 |
arl6
|
ADP-ribosylation factor-like 6 |
| chr20_+_41906960 | 0.04 |
ENSDART00000193460
|
cep85l
|
centrosomal protein 85, like |
| chr15_-_23783822 | 0.03 |
ENSDART00000180932
|
rad1
|
RAD1 homolog (S. pombe) |
| chr4_+_5868034 | 0.03 |
ENSDART00000166591
|
utp20
|
UTP20 small subunit (SSU) processome component |
| chr19_-_3240605 | 0.03 |
ENSDART00000105168
|
si:ch211-133n4.4
|
si:ch211-133n4.4 |
| chr5_+_20570573 | 0.03 |
ENSDART00000137019
|
cmklr1
|
chemokine-like receptor 1 |
| chr4_-_2168805 | 0.03 |
ENSDART00000150039
|
kcnc2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr20_-_34985555 | 0.03 |
ENSDART00000037065
|
sccpdhb
|
saccharopine dehydrogenase b |
| chr16_-_36798783 | 0.03 |
ENSDART00000145697
|
calb1
|
calbindin 1 |
| chr8_-_18460726 | 0.03 |
ENSDART00000149209
ENSDART00000129535 |
zgc:153738
|
zgc:153738 |
| chr6_+_30689239 | 0.03 |
ENSDART00000065206
|
wdr78
|
WD repeat domain 78 |
| chr12_-_8070969 | 0.03 |
ENSDART00000020995
|
tmem26b
|
transmembrane protein 26b |
| chr21_+_3093419 | 0.03 |
ENSDART00000162520
|
SHC3
|
SHC adaptor protein 3 |
| chr20_-_32752457 | 0.03 |
ENSDART00000153287
|
si:dkey-6f10.3
|
si:dkey-6f10.3 |
| chr12_+_28854963 | 0.03 |
ENSDART00000153227
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr10_-_32494304 | 0.03 |
ENSDART00000028161
|
uvrag
|
UV radiation resistance associated gene |
| chr4_-_16853464 | 0.03 |
ENSDART00000125743
ENSDART00000164570 |
slc25a3a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3a |
| chr12_+_34258139 | 0.03 |
ENSDART00000153127
|
socs3b
|
suppressor of cytokine signaling 3b |
| chr6_-_14040136 | 0.03 |
ENSDART00000065361
ENSDART00000179765 |
etv5b
|
ets variant 5b |
| chr8_-_18398787 | 0.03 |
ENSDART00000063577
|
obscna
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF a |
| chr5_-_26879302 | 0.03 |
ENSDART00000098571
ENSDART00000139086 |
zgc:64051
|
zgc:64051 |
| chr1_-_45752261 | 0.03 |
ENSDART00000141756
|
si:ch211-214c7.5
|
si:ch211-214c7.5 |
| chr22_+_16446052 | 0.03 |
ENSDART00000142454
|
si:dkey-121a11.3
|
si:dkey-121a11.3 |
| chr20_+_17739923 | 0.03 |
ENSDART00000024627
|
cdh2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr7_-_18168493 | 0.03 |
ENSDART00000127428
|
peli3
|
pellino E3 ubiquitin protein ligase family member 3 |
| chr16_+_42830152 | 0.03 |
ENSDART00000159730
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr15_-_20190052 | 0.03 |
ENSDART00000157149
|
exoc3l2b
|
exocyst complex component 3-like 2b |
| chr19_+_4059200 | 0.03 |
ENSDART00000161676
ENSDART00000172424 ENSDART00000161804 |
btr25
|
bloodthirsty-related gene family, member 25 |
| chr10_-_32494499 | 0.03 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr5_-_18046053 | 0.03 |
ENSDART00000144898
|
rnf215
|
ring finger protein 215 |
| chr23_+_25354856 | 0.03 |
ENSDART00000109023
ENSDART00000147440 |
fmnl3
|
formin-like 3 |
| chr3_-_22366562 | 0.03 |
ENSDART00000129447
|
ifnphi1
|
interferon phi 1 |
| chr21_+_6613772 | 0.03 |
ENSDART00000159645
|
col5a1
|
procollagen, type V, alpha 1 |
| chr2_-_1514001 | 0.03 |
ENSDART00000057736
|
c8b
|
complement component 8, beta polypeptide |
| chr23_+_19213472 | 0.03 |
ENSDART00000185985
|
apobec2b
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2b |
| chr17_-_46990003 | 0.03 |
ENSDART00000155257
|
si:ch211-244k5.1
|
si:ch211-244k5.1 |
| chr3_-_25148047 | 0.03 |
ENSDART00000089325
|
mief1
|
mitochondrial elongation factor 1 |
| chr12_+_23912074 | 0.02 |
ENSDART00000152864
|
svila
|
supervillin a |
| chr8_-_11546175 | 0.02 |
ENSDART00000081909
|
si:ch211-248e11.2
|
si:ch211-248e11.2 |
| chr23_+_32406009 | 0.02 |
ENSDART00000155793
|
si:ch211-66i15.5
|
si:ch211-66i15.5 |
| chr9_+_51225345 | 0.02 |
ENSDART00000132896
|
fap
|
fibroblast activation protein, alpha |
| chr11_-_16215143 | 0.02 |
ENSDART00000027014
|
rab7
|
RAB7, member RAS oncogene family |
| chr11_+_36665359 | 0.02 |
ENSDART00000166144
|
si:ch211-11c3.9
|
si:ch211-11c3.9 |
| chr16_+_9540033 | 0.02 |
ENSDART00000149574
|
ca14
|
carbonic anhydrase XIV |
| chr8_-_1956632 | 0.02 |
ENSDART00000131602
|
si:dkey-178e17.1
|
si:dkey-178e17.1 |
| chr4_-_1839352 | 0.02 |
ENSDART00000189215
|
pwp1
|
PWP1 homolog (S. cerevisiae) |
| chr20_+_700616 | 0.02 |
ENSDART00000168166
|
senp6a
|
SUMO1/sentrin specific peptidase 6a |
| chr2_-_9059955 | 0.02 |
ENSDART00000022768
|
ak5
|
adenylate kinase 5 |
| chr18_-_34549721 | 0.02 |
ENSDART00000137101
ENSDART00000021880 |
ssr3
|
signal sequence receptor, gamma |
| chr11_-_12673637 | 0.02 |
ENSDART00000186058
|
CR450764.7
|
|
| chr18_+_8833251 | 0.02 |
ENSDART00000143519
|
impdh1a
|
IMP (inosine 5'-monophosphate) dehydrogenase 1a |
| chr4_+_57101749 | 0.02 |
ENSDART00000135121
|
si:ch211-238e22.4
|
si:ch211-238e22.4 |
| chr9_+_42095220 | 0.02 |
ENSDART00000148317
ENSDART00000134431 |
pcbp3
|
poly(rC) binding protein 3 |
| chr19_-_3127271 | 0.02 |
ENSDART00000146928
ENSDART00000134197 |
si:ch211-80h18.1
|
si:ch211-80h18.1 |
| chr14_-_7045009 | 0.02 |
ENSDART00000112082
|
rufy1
|
RUN and FYVE domain containing 1 |
| chr16_+_20947439 | 0.02 |
ENSDART00000137344
|
skap2
|
src kinase associated phosphoprotein 2 |
| chr13_+_17672527 | 0.02 |
ENSDART00000148269
ENSDART00000137776 |
comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr4_-_1839192 | 0.02 |
ENSDART00000003790
|
pwp1
|
PWP1 homolog (S. cerevisiae) |
| chr16_-_38629208 | 0.02 |
ENSDART00000126705
|
eif3ea
|
eukaryotic translation initiation factor 3, subunit E, a |
| chr16_+_27593698 | 0.02 |
ENSDART00000145203
|
si:ch211-197h24.8
|
si:ch211-197h24.8 |
| chr23_-_15916316 | 0.02 |
ENSDART00000134096
ENSDART00000042469 ENSDART00000146605 |
mrgbp
|
MRG/MORF4L binding protein |
| chr2_-_55298075 | 0.02 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr16_+_50972803 | 0.02 |
ENSDART00000178189
|
si:dkeyp-97a10.2
|
si:dkeyp-97a10.2 |
| chr4_+_45148652 | 0.02 |
ENSDART00000150798
|
si:dkey-51d8.9
|
si:dkey-51d8.9 |
| chr8_-_32354677 | 0.02 |
ENSDART00000138268
ENSDART00000133245 ENSDART00000179677 ENSDART00000174450 |
ipo11
|
importin 11 |
| chr10_-_5053589 | 0.02 |
ENSDART00000193579
|
tmem150c
|
transmembrane protein 150C |
| chr2_-_59145027 | 0.02 |
ENSDART00000128320
|
FO834803.1
|
|
| chr9_-_7378566 | 0.02 |
ENSDART00000144003
|
slc23a3
|
solute carrier family 23, member 3 |
| chr11_-_16215470 | 0.02 |
ENSDART00000185744
|
rab7
|
RAB7, member RAS oncogene family |
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb16a+zbtb16b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.2 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.0 | 0.2 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0016093 | polyprenol metabolic process(GO:0016093) |
| 0.0 | 0.1 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.0 | 0.1 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.0 | 0.1 | GO:0018063 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 0.3 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.4 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 0.1 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.0 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.0 | 0.0 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.1 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.1 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.1 | GO:0030329 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0071539 | protein localization to centrosome(GO:0071539) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.4 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.2 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.0 | 0.1 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0052855 | ATP-dependent NAD(P)H-hydrate dehydratase activity(GO:0047453) ADP-dependent NAD(P)H-hydrate dehydratase activity(GO:0052855) |
| 0.0 | 0.1 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.0 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.0 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.0 | 0.1 | GO:0005537 | mannose binding(GO:0005537) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |