Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
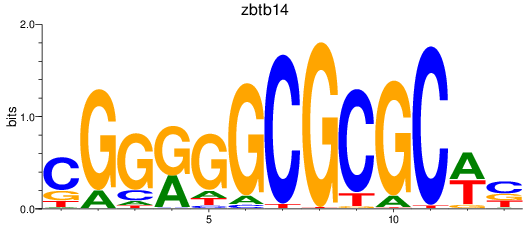
Results for zbtb14
Z-value: 0.40
Transcription factors associated with zbtb14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zbtb14
|
ENSDARG00000098273 | zinc finger and BTB domain containing 14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zbtb14 | dr11_v1_chr24_+_41989108_41989108 | -0.23 | 3.6e-01 | Click! |
Activity profile of zbtb14 motif
Sorted Z-values of zbtb14 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_25350435 | 0.91 |
ENSDART00000063034
|
fam228a
|
family with sequence similarity 228, member A |
| chr7_+_73332486 | 0.74 |
ENSDART00000174119
ENSDART00000092051 ENSDART00000192388 |
CABZ01081780.1
|
|
| chr24_+_32176155 | 0.73 |
ENSDART00000003745
|
vim
|
vimentin |
| chr14_+_46313135 | 0.65 |
ENSDART00000172902
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr3_-_31784082 | 0.63 |
ENSDART00000134201
|
kcnh6a
|
potassium voltage-gated channel, subfamily H (eag-related), member 6a |
| chr12_-_7607114 | 0.61 |
ENSDART00000158095
|
slc16a9b
|
solute carrier family 16, member 9b |
| chr1_-_53700392 | 0.61 |
ENSDART00000127216
|
fam161a
|
family with sequence similarity 161, member A |
| chr7_+_67451108 | 0.59 |
ENSDART00000163840
|
gcshb
|
glycine cleavage system protein H (aminomethyl carrier), b |
| chr13_+_111212 | 0.55 |
ENSDART00000167840
|
dnaaf2
|
dynein, axonemal, assembly factor 2 |
| chr21_-_8422351 | 0.50 |
ENSDART00000055329
ENSDART00000134360 |
lhx2a
|
LIM homeobox 2a |
| chr18_+_44795711 | 0.46 |
ENSDART00000110229
ENSDART00000188262 ENSDART00000139526 |
fam118b
|
family with sequence similarity 118, member B |
| chr24_-_28245872 | 0.46 |
ENSDART00000167861
|
cox4i1l
|
cytochrome c oxidase subunit IV isoform 1, like |
| chr11_+_41858807 | 0.46 |
ENSDART00000161605
|
iffo2b
|
intermediate filament family orphan 2b |
| chr19_+_3653976 | 0.45 |
ENSDART00000125673
|
nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr24_+_5840748 | 0.44 |
ENSDART00000139191
|
trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr1_-_56223913 | 0.44 |
ENSDART00000019573
|
zgc:65894
|
zgc:65894 |
| chr4_-_797831 | 0.42 |
ENSDART00000158970
ENSDART00000170012 |
mapre3b
|
microtubule-associated protein, RP/EB family, member 3b |
| chr24_+_5840258 | 0.40 |
ENSDART00000087034
|
trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr4_+_22680442 | 0.40 |
ENSDART00000036531
|
gnai1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr6_+_11438972 | 0.39 |
ENSDART00000029314
|
col5a2b
|
collagen, type V, alpha 2b |
| chr4_-_22472653 | 0.39 |
ENSDART00000066903
ENSDART00000130072 ENSDART00000123369 |
kmt2e
|
lysine (K)-specific methyltransferase 2E |
| chr15_+_36941490 | 0.38 |
ENSDART00000172664
|
kirrel3l
|
kirre like nephrin family adhesion molecule 3, like |
| chr5_-_10002260 | 0.38 |
ENSDART00000141831
|
si:ch73-266o15.4
|
si:ch73-266o15.4 |
| chr25_-_3979583 | 0.38 |
ENSDART00000124749
|
myrf
|
myelin regulatory factor |
| chr5_-_34185497 | 0.37 |
ENSDART00000146321
|
fibcd1
|
fibrinogen C domain containing 1 |
| chr12_-_1951233 | 0.37 |
ENSDART00000005676
ENSDART00000127937 |
sox9a
|
SRY (sex determining region Y)-box 9a |
| chr10_-_27223827 | 0.35 |
ENSDART00000185138
|
auts2a
|
autism susceptibility candidate 2a |
| chr23_+_44644911 | 0.35 |
ENSDART00000140799
|
zgc:85858
|
zgc:85858 |
| chr10_-_690072 | 0.35 |
ENSDART00000164871
ENSDART00000142833 |
glis3
|
GLIS family zinc finger 3 |
| chr21_+_11916788 | 0.35 |
ENSDART00000136103
|
ubap2a
|
ubiquitin associated protein 2a |
| chr3_+_36313532 | 0.34 |
ENSDART00000151305
|
slc16a6b
|
solute carrier family 16, member 6b |
| chr6_+_120181 | 0.34 |
ENSDART00000151209
ENSDART00000185930 |
cdkn2d
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr25_+_14165447 | 0.33 |
ENSDART00000145387
|
shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr20_+_23440632 | 0.33 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr23_-_44226556 | 0.32 |
ENSDART00000149115
|
zgc:158659
|
zgc:158659 |
| chr14_+_46313396 | 0.31 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr4_+_306036 | 0.31 |
ENSDART00000103659
|
msgn1
|
mesogenin 1 |
| chr22_-_13042992 | 0.31 |
ENSDART00000028787
|
ahr1b
|
aryl hydrocarbon receptor 1b |
| chr20_+_18209895 | 0.30 |
ENSDART00000111063
|
kctd1
|
potassium channel tetramerization domain containing 1 |
| chr22_+_38978084 | 0.30 |
ENSDART00000025482
|
arhgef3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr22_-_21314821 | 0.30 |
ENSDART00000105546
ENSDART00000135388 |
cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr23_-_306796 | 0.29 |
ENSDART00000143125
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr11_-_6206520 | 0.29 |
ENSDART00000150199
ENSDART00000148246 ENSDART00000019440 |
pole4
|
polymerase (DNA-directed), epsilon 4, accessory subunit |
| chr16_+_54248706 | 0.29 |
ENSDART00000156915
|
drd2l
|
dopamine receptor D2 like |
| chr8_-_29719393 | 0.29 |
ENSDART00000077635
|
si:ch211-103n10.5
|
si:ch211-103n10.5 |
| chr25_+_37209619 | 0.27 |
ENSDART00000112192
|
si:dkey-234i14.3
|
si:dkey-234i14.3 |
| chr16_+_28754403 | 0.26 |
ENSDART00000103340
|
s100v1
|
S100 calcium binding protein V1 |
| chr6_+_46258866 | 0.26 |
ENSDART00000134584
|
zgc:162324
|
zgc:162324 |
| chr16_-_13388821 | 0.26 |
ENSDART00000144062
|
grin2db
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, b |
| chr18_-_44527124 | 0.26 |
ENSDART00000189471
|
aplp2
|
amyloid beta (A4) precursor-like protein 2 |
| chr25_-_3979288 | 0.25 |
ENSDART00000157117
|
myrf
|
myelin regulatory factor |
| chr3_-_6441619 | 0.24 |
ENSDART00000157771
ENSDART00000166758 |
sumo2b
|
small ubiquitin-like modifier 2b |
| chr18_-_44526940 | 0.24 |
ENSDART00000077125
|
aplp2
|
amyloid beta (A4) precursor-like protein 2 |
| chr2_-_22688651 | 0.24 |
ENSDART00000013863
|
agxtb
|
alanine-glyoxylate aminotransferase b |
| chr23_-_20258490 | 0.23 |
ENSDART00000147326
|
lamb2
|
laminin, beta 2 (laminin S) |
| chr20_-_40451115 | 0.23 |
ENSDART00000075092
|
pkib
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr15_-_47929455 | 0.22 |
ENSDART00000064462
|
psma6l
|
proteasome subunit alpha 6, like |
| chr3_+_39099716 | 0.22 |
ENSDART00000083388
|
tmem98
|
transmembrane protein 98 |
| chr19_-_13733870 | 0.22 |
ENSDART00000177773
|
epb41a
|
erythrocyte membrane protein band 4.1a |
| chr24_+_10413484 | 0.21 |
ENSDART00000111014
|
myca
|
MYC proto-oncogene, bHLH transcription factor a |
| chr15_-_47822597 | 0.21 |
ENSDART00000193236
ENSDART00000161391 |
CZQB01095947.1
|
|
| chr17_+_53311618 | 0.20 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr2_+_2918759 | 0.20 |
ENSDART00000161255
|
CABZ01112864.1
|
|
| chr12_-_30032188 | 0.20 |
ENSDART00000042514
|
atrnl1b
|
attractin-like 1b |
| chr9_-_44289636 | 0.20 |
ENSDART00000110411
|
cerkl
|
ceramide kinase-like |
| chr18_+_6155258 | 0.20 |
ENSDART00000092804
|
otogl
|
otogelin-like |
| chr4_-_9810999 | 0.20 |
ENSDART00000146858
|
si:dkeyp-27e10.3
|
si:dkeyp-27e10.3 |
| chr1_-_46984142 | 0.20 |
ENSDART00000125032
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr25_-_3644574 | 0.19 |
ENSDART00000188569
|
si:ch211-272n13.3
|
si:ch211-272n13.3 |
| chr4_+_6833583 | 0.19 |
ENSDART00000165179
ENSDART00000186134 ENSDART00000174507 |
dock4b
|
dedicator of cytokinesis 4b |
| chr19_+_41075488 | 0.19 |
ENSDART00000138687
|
ppp1r9a
|
protein phosphatase 1, regulatory subunit 9A |
| chr13_-_40401870 | 0.19 |
ENSDART00000128951
|
nkx3.3
|
NK3 homeobox 3 |
| chr22_-_12160283 | 0.19 |
ENSDART00000146785
ENSDART00000128176 |
tmem163b
|
transmembrane protein 163b |
| chr1_-_38709551 | 0.19 |
ENSDART00000128794
|
gpm6ab
|
glycoprotein M6Ab |
| chr7_+_30926289 | 0.19 |
ENSDART00000173518
|
si:dkey-1h4.4
|
si:dkey-1h4.4 |
| chr25_-_18140537 | 0.19 |
ENSDART00000113581
|
kitlga
|
kit ligand a |
| chr13_-_31470439 | 0.18 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr5_-_67661102 | 0.18 |
ENSDART00000013605
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr9_-_46842179 | 0.18 |
ENSDART00000054137
|
igfbp5b
|
insulin-like growth factor binding protein 5b |
| chr3_-_62087346 | 0.18 |
ENSDART00000092665
|
srebf1
|
sterol regulatory element binding transcription factor 1 |
| chr24_-_35699595 | 0.18 |
ENSDART00000167990
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr17_-_43666166 | 0.18 |
ENSDART00000077990
|
egr2a
|
early growth response 2a |
| chr9_-_55128839 | 0.18 |
ENSDART00000085135
|
tbl1x
|
transducin beta like 1 X-linked |
| chr5_+_63668735 | 0.18 |
ENSDART00000134261
ENSDART00000097330 |
dnm1b
|
dynamin 1b |
| chr9_-_52814204 | 0.18 |
ENSDART00000140771
ENSDART00000007401 |
MAP3K13
|
si:ch211-45c16.2 |
| chr19_+_41551543 | 0.18 |
ENSDART00000112364
|
si:ch211-57n23.4
|
si:ch211-57n23.4 |
| chr16_+_11242443 | 0.18 |
ENSDART00000024935
|
gsk3ab
|
glycogen synthase kinase 3 alpha b |
| chr24_-_1023112 | 0.18 |
ENSDART00000147508
ENSDART00000066863 |
ralaa
|
v-ral simian leukemia viral oncogene homolog Aa (ras related) |
| chr12_+_33894665 | 0.17 |
ENSDART00000004769
|
mfsd13a
|
major facilitator superfamily domain containing 13A |
| chr22_-_7006974 | 0.17 |
ENSDART00000133143
|
gpd1b
|
glycerol-3-phosphate dehydrogenase 1b |
| chr20_-_2725594 | 0.17 |
ENSDART00000152120
|
akirin2
|
akirin 2 |
| chr3_-_1204341 | 0.17 |
ENSDART00000089646
|
fam234b
|
family with sequence similarity 234, member B |
| chr1_-_36151377 | 0.17 |
ENSDART00000037516
|
znf827
|
zinc finger protein 827 |
| chr17_+_51764310 | 0.17 |
ENSDART00000157171
|
si:ch211-168d23.3
|
si:ch211-168d23.3 |
| chr22_+_3045495 | 0.16 |
ENSDART00000164061
|
LO017843.1
|
|
| chr24_-_35699444 | 0.16 |
ENSDART00000166567
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr20_-_3953631 | 0.16 |
ENSDART00000124197
|
CU929133.1
|
|
| chr7_-_37858569 | 0.16 |
ENSDART00000129156
ENSDART00000173552 ENSDART00000178306 |
adcy7
|
adenylate cyclase 7 |
| chr1_+_40741198 | 0.16 |
ENSDART00000147672
ENSDART00000163584 |
htra3a
|
HtrA serine peptidase 3a |
| chr13_+_35746440 | 0.16 |
ENSDART00000187859
|
gpr75
|
G protein-coupled receptor 75 |
| chr16_-_43233509 | 0.16 |
ENSDART00000025877
|
cldn12
|
claudin 12 |
| chr2_+_58841181 | 0.16 |
ENSDART00000164102
|
cirbpa
|
cold inducible RNA binding protein a |
| chr10_-_25069155 | 0.16 |
ENSDART00000078226
ENSDART00000181941 |
mtnr1bb
|
melatonin receptor 1Bb |
| chr24_+_17068724 | 0.15 |
ENSDART00000191137
|
pip4k2aa
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha a |
| chr2_-_32386791 | 0.15 |
ENSDART00000056634
|
ubtfl
|
upstream binding transcription factor, like |
| chr11_-_24347644 | 0.15 |
ENSDART00000089777
|
si:ch211-15p9.2
|
si:ch211-15p9.2 |
| chr10_+_6121558 | 0.15 |
ENSDART00000166799
ENSDART00000157947 |
tln1
|
talin 1 |
| chr16_+_3004422 | 0.15 |
ENSDART00000189969
|
CABZ01043952.1
|
|
| chr20_-_31067306 | 0.15 |
ENSDART00000014163
|
fndc1
|
fibronectin type III domain containing 1 |
| chr14_-_970853 | 0.15 |
ENSDART00000130801
|
acsl1b
|
acyl-CoA synthetase long chain family member 1b |
| chr4_+_9028819 | 0.15 |
ENSDART00000102893
|
aldh1l2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr6_+_52350443 | 0.15 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr11_-_45385803 | 0.14 |
ENSDART00000173329
|
trappc10
|
trafficking protein particle complex 10 |
| chr25_-_15040369 | 0.14 |
ENSDART00000159342
ENSDART00000166490 |
pax6a
|
paired box 6a |
| chr3_-_4501026 | 0.14 |
ENSDART00000163052
|
zgc:162198
|
zgc:162198 |
| chr2_+_38742338 | 0.14 |
ENSDART00000177290
|
carmil3
|
capping protein regulator and myosin 1 linker 3 |
| chr20_-_1314537 | 0.14 |
ENSDART00000144288
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr12_-_35885349 | 0.14 |
ENSDART00000085662
|
cep131
|
centrosomal protein 131 |
| chr21_-_25573064 | 0.14 |
ENSDART00000134310
|
CR388166.1
|
|
| chr3_-_31845816 | 0.14 |
ENSDART00000157028
|
map3k3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr16_-_45288599 | 0.14 |
ENSDART00000044097
|
hpn
|
hepsin |
| chr25_+_3677650 | 0.14 |
ENSDART00000154348
|
prnprs3
|
prion protein, related sequence 3 |
| chr2_+_36046126 | 0.14 |
ENSDART00000085976
ENSDART00000171680 |
smg7
|
SMG7 nonsense mediated mRNA decay factor |
| chr3_-_18575868 | 0.14 |
ENSDART00000122968
|
aqp8b
|
aquaporin 8b |
| chr2_+_45532437 | 0.14 |
ENSDART00000137465
|
aknad1
|
AKNA domain containing 1 |
| chr20_-_54869006 | 0.14 |
ENSDART00000184817
|
CABZ01037174.1
|
|
| chr1_-_8428736 | 0.13 |
ENSDART00000138435
ENSDART00000121823 |
syngr3b
|
synaptogyrin 3b |
| chr11_-_45385602 | 0.13 |
ENSDART00000166691
|
trappc10
|
trafficking protein particle complex 10 |
| chr6_+_48862 | 0.13 |
ENSDART00000082954
|
mbd5
|
methyl-CpG binding domain protein 5 |
| chr7_-_24236364 | 0.13 |
ENSDART00000010124
|
slc7a8a
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8a |
| chr17_-_51829310 | 0.13 |
ENSDART00000154544
|
numb
|
numb homolog (Drosophila) |
| chr25_+_4837915 | 0.13 |
ENSDART00000168016
|
gnb5a
|
guanine nucleotide binding protein (G protein), beta 5a |
| chr22_-_22416337 | 0.13 |
ENSDART00000142947
ENSDART00000089569 |
camsap2a
|
calmodulin regulated spectrin-associated protein family, member 2a |
| chr22_+_12431608 | 0.13 |
ENSDART00000108609
|
rnd3a
|
Rho family GTPase 3a |
| chr3_-_44012748 | 0.12 |
ENSDART00000167248
ENSDART00000157463 ENSDART00000159111 |
il4r.1
|
interleukin 4 receptor, tandem duplicate 1 |
| chr19_+_25649626 | 0.12 |
ENSDART00000146947
|
tac1
|
tachykinin 1 |
| chr19_+_41551335 | 0.12 |
ENSDART00000169193
|
si:ch211-57n23.4
|
si:ch211-57n23.4 |
| chr4_+_357810 | 0.12 |
ENSDART00000163436
ENSDART00000103645 |
tmem181
|
transmembrane protein 181 |
| chr4_+_9177997 | 0.12 |
ENSDART00000057254
ENSDART00000154614 |
nfyba
|
nuclear transcription factor Y, beta a |
| chr14_-_8795081 | 0.12 |
ENSDART00000106671
|
tgfb2l
|
transforming growth factor, beta 2, like |
| chr23_-_31400792 | 0.12 |
ENSDART00000132736
|
lca5
|
Leber congenital amaurosis 5 |
| chr20_-_26531850 | 0.12 |
ENSDART00000183317
ENSDART00000131994 |
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr25_+_7346800 | 0.12 |
ENSDART00000154404
|
peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr2_-_30324297 | 0.12 |
ENSDART00000099078
|
jph1b
|
junctophilin 1b |
| chr1_+_54115839 | 0.12 |
ENSDART00000180214
|
LO017722.2
|
|
| chr20_+_46311707 | 0.12 |
ENSDART00000184743
|
flvcr2b
|
feline leukemia virus subgroup C cellular receptor family, member 2b |
| chr25_-_35121946 | 0.11 |
ENSDART00000137502
|
si:ch73-36p18.1
|
si:ch73-36p18.1 |
| chr3_+_32789605 | 0.11 |
ENSDART00000171895
|
tbc1d10b
|
TBC1 domain family, member 10b |
| chr25_+_35061044 | 0.11 |
ENSDART00000111706
|
si:dkey-108k21.14
|
si:dkey-108k21.14 |
| chr4_-_12007404 | 0.11 |
ENSDART00000092250
|
btbd11a
|
BTB (POZ) domain containing 11a |
| chr20_+_54666222 | 0.11 |
ENSDART00000166592
|
CABZ01087948.1
|
|
| chr4_-_2350371 | 0.11 |
ENSDART00000166274
|
phlda1
|
pleckstrin homology-like domain, family A, member 1 |
| chr5_-_64454459 | 0.11 |
ENSDART00000172321
ENSDART00000168030 |
brd3b
|
bromodomain containing 3b |
| chr12_+_47698356 | 0.11 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr15_-_29354020 | 0.11 |
ENSDART00000127795
|
tsku
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis) |
| chr9_-_14504834 | 0.11 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr13_-_5569562 | 0.11 |
ENSDART00000102576
|
meis1b
|
Meis homeobox 1 b |
| chr9_+_44832032 | 0.11 |
ENSDART00000002633
|
frzb
|
frizzled related protein |
| chr18_-_5692292 | 0.11 |
ENSDART00000121503
|
cplx3b
|
complexin 3b |
| chr4_-_779796 | 0.11 |
ENSDART00000128743
|
tmem214
|
transmembrane protein 214 |
| chr11_-_33960318 | 0.11 |
ENSDART00000087597
|
col6a2
|
collagen, type VI, alpha 2 |
| chr2_-_24462277 | 0.11 |
ENSDART00000033922
|
kcnn1a
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1a |
| chr1_-_50859053 | 0.11 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr5_-_44496805 | 0.10 |
ENSDART00000110076
|
gas1a
|
growth arrest-specific 1a |
| chr18_+_41820039 | 0.10 |
ENSDART00000128345
|
pgr
|
progesterone receptor |
| chr4_+_6833735 | 0.10 |
ENSDART00000136355
|
dock4b
|
dedicator of cytokinesis 4b |
| chr11_-_45420212 | 0.10 |
ENSDART00000182042
ENSDART00000163185 |
ankrd13c
|
ankyrin repeat domain 13C |
| chr7_+_73593814 | 0.10 |
ENSDART00000110544
|
znf219
|
zinc finger protein 219 |
| chr2_-_44720551 | 0.10 |
ENSDART00000146380
|
map6d1
|
MAP6 domain containing 1 |
| chr18_+_62932 | 0.10 |
ENSDART00000052638
|
slc27a2a
|
solute carrier family 27 (fatty acid transporter), member 2a |
| chr16_+_45456066 | 0.10 |
ENSDART00000093365
|
syngap1b
|
synaptic Ras GTPase activating protein 1b |
| chr20_-_23440955 | 0.10 |
ENSDART00000153386
|
slc10a4
|
solute carrier family 10, member 4 |
| chr1_+_53321878 | 0.10 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr5_-_10239079 | 0.10 |
ENSDART00000132739
|
si:ch73-42k18.1
|
si:ch73-42k18.1 |
| chr3_-_21348478 | 0.10 |
ENSDART00000114906
|
fam171a2a
|
family with sequence similarity 171, member A2a |
| chr2_-_32386598 | 0.10 |
ENSDART00000145575
|
ubtfl
|
upstream binding transcription factor, like |
| chr20_-_20312789 | 0.10 |
ENSDART00000114779
|
si:ch211-212g7.6
|
si:ch211-212g7.6 |
| chr12_+_33894396 | 0.10 |
ENSDART00000130853
ENSDART00000152988 |
mfsd13a
|
major facilitator superfamily domain containing 13A |
| chr21_+_6212844 | 0.10 |
ENSDART00000150301
|
fnbp1b
|
formin binding protein 1b |
| chr20_-_35246150 | 0.10 |
ENSDART00000090549
|
fzd3a
|
frizzled class receptor 3a |
| chr10_+_17592273 | 0.10 |
ENSDART00000141221
|
KCNN2
|
si:dkey-76p7.5 |
| chr25_-_25550938 | 0.09 |
ENSDART00000150412
ENSDART00000103622 |
irf7
|
interferon regulatory factor 7 |
| chr13_+_18471546 | 0.09 |
ENSDART00000090712
ENSDART00000127962 |
stox1
|
storkhead box 1 |
| chr20_-_26532167 | 0.09 |
ENSDART00000061914
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr15_-_23761580 | 0.09 |
ENSDART00000137918
|
bbc3
|
BCL2 binding component 3 |
| chr7_+_1383027 | 0.09 |
ENSDART00000143762
|
khnyn
|
KH and NYN domain containing |
| chr11_+_44932852 | 0.09 |
ENSDART00000172840
|
si:cabz01080295.1
|
si:cabz01080295.1 |
| chr20_-_1314355 | 0.09 |
ENSDART00000152436
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr1_-_20911297 | 0.09 |
ENSDART00000078271
|
cpe
|
carboxypeptidase E |
| chr16_-_42152145 | 0.09 |
ENSDART00000038748
|
dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr4_+_77933084 | 0.09 |
ENSDART00000148728
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr11_+_42641404 | 0.09 |
ENSDART00000172641
ENSDART00000169938 |
il17rd
|
interleukin 17 receptor D |
| chr22_+_1170294 | 0.09 |
ENSDART00000159761
ENSDART00000169809 |
irf6
|
interferon regulatory factor 6 |
| chr10_+_25204626 | 0.09 |
ENSDART00000024514
|
chordc1b
|
cysteine and histidine-rich domain (CHORD) containing 1b |
| chr12_-_28363111 | 0.08 |
ENSDART00000016283
ENSDART00000164156 |
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr11_-_10828539 | 0.08 |
ENSDART00000040180
|
tbr1a
|
T-box, brain, 1a |
| chr16_-_12784373 | 0.08 |
ENSDART00000080396
|
foxj2
|
forkhead box J2 |
| chr20_-_44557037 | 0.08 |
ENSDART00000140995
|
mfsd2b
|
major facilitator superfamily domain containing 2B |
| chr18_-_50799510 | 0.08 |
ENSDART00000174373
|
taldo1
|
transaldolase 1 |
| chr23_+_41800052 | 0.08 |
ENSDART00000141484
|
pdyn
|
prodynorphin |
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb14
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0099625 | regulation of membrane repolarization(GO:0060306) regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.1 | 0.6 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.3 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.1 | 0.3 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.5 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.1 | 0.5 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.8 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.6 | GO:0043217 | myelin maintenance(GO:0043217) |
| 0.1 | 0.2 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) regulation of apoptotic cell clearance(GO:2000425) |
| 0.0 | 0.2 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.1 | GO:0009397 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.0 | 0.1 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.0 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.2 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.2 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:1904184 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.0 | 0.2 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.0 | 0.1 | GO:0097037 | heme export(GO:0097037) |
| 0.0 | 0.1 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 | 0.1 | GO:0021731 | trigeminal motor nucleus development(GO:0021731) |
| 0.0 | 0.1 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.0 | 0.5 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 1.1 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.4 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.0 | 0.3 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.5 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.1 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.1 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.0 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.4 | GO:0039022 | pronephric duct development(GO:0039022) |
| 0.0 | 0.1 | GO:0032608 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0090199 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.0 | 0.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.0 | 0.0 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.1 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.1 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.0 | 0.1 | GO:0048913 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.0 | 0.4 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.1 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.3 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.1 | GO:0070316 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 0.5 | GO:0051057 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 0.1 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 0.3 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0034451 | acrosomal vesicle(GO:0001669) centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.2 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0031835 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.0 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.2 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.4 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.1 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.5 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.9 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.0 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.6 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |