Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
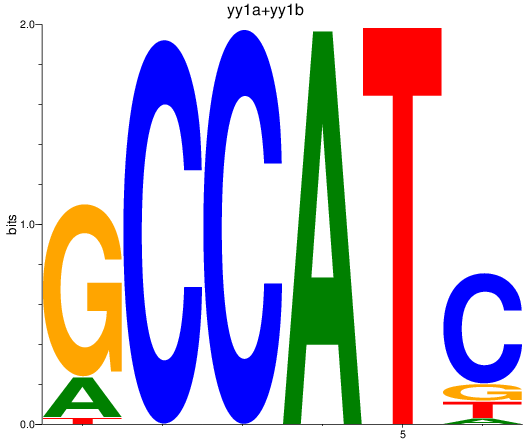
Results for yy1a+yy1b
Z-value: 1.91
Transcription factors associated with yy1a+yy1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
yy1b
|
ENSDARG00000027978 | YY1 transcription factor b |
|
yy1a
|
ENSDARG00000042796 | YY1 transcription factor a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| yy1a | dr11_v1_chr17_-_30863252_30863252 | -0.74 | 5.1e-04 | Click! |
| yy1b | dr11_v1_chr20_-_54435287_54435287 | -0.64 | 4.3e-03 | Click! |
Activity profile of yy1a+yy1b motif
Sorted Z-values of yy1a+yy1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_41468892 | 5.44 |
ENSDART00000173099
ENSDART00000003170 |
mid1ip1l
|
MID1 interacting protein 1, like |
| chr1_-_18811517 | 4.97 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr23_-_44577885 | 4.59 |
ENSDART00000166654
|
si:ch73-160p18.4
|
si:ch73-160p18.4 |
| chr15_-_35126332 | 4.34 |
ENSDART00000007636
|
zgc:55413
|
zgc:55413 |
| chr11_+_14333441 | 3.94 |
ENSDART00000171969
|
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr2_+_68789 | 3.67 |
ENSDART00000058569
|
cldn1
|
claudin 1 |
| chr9_+_2393764 | 3.67 |
ENSDART00000172624
|
chn1
|
chimerin 1 |
| chr11_+_18053333 | 3.65 |
ENSDART00000075750
|
zgc:175135
|
zgc:175135 |
| chr11_+_18130300 | 3.63 |
ENSDART00000169146
|
zgc:175135
|
zgc:175135 |
| chr21_-_13690712 | 3.53 |
ENSDART00000065817
|
pou5f3
|
POU domain, class 5, transcription factor 3 |
| chr7_+_27603211 | 3.46 |
ENSDART00000148782
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr1_+_218524 | 3.38 |
ENSDART00000109529
|
tmco3
|
transmembrane and coiled-coil domains 3 |
| chr12_+_31783066 | 3.35 |
ENSDART00000105584
|
lrrc59
|
leucine rich repeat containing 59 |
| chr14_-_35892767 | 3.34 |
ENSDART00000052648
|
tmem144b
|
transmembrane protein 144b |
| chr8_+_3434146 | 3.25 |
ENSDART00000164426
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr11_+_18157260 | 3.19 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr11_+_18037729 | 3.17 |
ENSDART00000111624
|
zgc:175135
|
zgc:175135 |
| chr9_+_8396755 | 3.09 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr7_-_4107423 | 3.07 |
ENSDART00000172971
|
zgc:55733
|
zgc:55733 |
| chr16_-_8120203 | 3.04 |
ENSDART00000193430
|
snrka
|
SNF related kinase a |
| chr5_+_3891485 | 3.01 |
ENSDART00000129329
ENSDART00000091711 |
rpain
|
RPA interacting protein |
| chr20_-_30035326 | 2.99 |
ENSDART00000141068
|
sox11b
|
SRY (sex determining region Y)-box 11b |
| chr6_-_8360918 | 2.78 |
ENSDART00000004716
|
acp5a
|
acid phosphatase 5a, tartrate resistant |
| chr23_+_7692042 | 2.78 |
ENSDART00000018512
|
pofut1
|
protein O-fucosyltransferase 1 |
| chr10_-_34002185 | 2.71 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr3_+_43102010 | 2.70 |
ENSDART00000162096
|
micall2a
|
mical-like 2a |
| chr8_-_12847483 | 2.69 |
ENSDART00000146186
|
si:dkey-104n9.1
|
si:dkey-104n9.1 |
| chr22_+_25184459 | 2.67 |
ENSDART00000105308
|
si:ch211-226h8.4
|
si:ch211-226h8.4 |
| chr18_-_46183462 | 2.64 |
ENSDART00000021192
|
kcnk6
|
potassium channel, subfamily K, member 6 |
| chr17_-_25331439 | 2.63 |
ENSDART00000155422
ENSDART00000082324 |
zpcx
|
zona pellucida protein C |
| chr12_+_20693743 | 2.58 |
ENSDART00000153023
ENSDART00000153370 |
st6galnac1.2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1, tandem duplicate 2 |
| chr8_+_1009831 | 2.46 |
ENSDART00000172414
|
fabp1b.2
|
fatty acid binding protein 1b, liver, tandem duplicate 2 |
| chr10_+_14982977 | 2.42 |
ENSDART00000140869
|
si:dkey-88l16.3
|
si:dkey-88l16.3 |
| chr15_+_24005289 | 2.40 |
ENSDART00000088648
|
myo18ab
|
myosin XVIIIAb |
| chr10_+_15608326 | 2.38 |
ENSDART00000188770
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr5_-_9678075 | 2.38 |
ENSDART00000097217
|
FAM109B
|
si:ch211-193c2.2 |
| chr14_-_45551572 | 2.38 |
ENSDART00000111410
|
ganab
|
glucosidase, alpha; neutral AB |
| chr10_+_3145707 | 2.37 |
ENSDART00000160046
|
hic2
|
hypermethylated in cancer 2 |
| chr22_+_14117078 | 2.35 |
ENSDART00000013575
|
bzw1a
|
basic leucine zipper and W2 domains 1a |
| chr20_-_43741159 | 2.35 |
ENSDART00000192621
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr8_+_36503797 | 2.34 |
ENSDART00000184785
|
slc7a4
|
solute carrier family 7, member 4 |
| chr22_+_5120033 | 2.32 |
ENSDART00000169200
|
mibp
|
muscle-specific beta 1 integrin binding protein |
| chr17_+_33158350 | 2.30 |
ENSDART00000104476
|
snx9a
|
sorting nexin 9a |
| chr7_+_1473929 | 2.29 |
ENSDART00000050687
|
lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr22_+_24318131 | 2.29 |
ENSDART00000187360
ENSDART00000165618 |
ccdc50
|
coiled-coil domain containing 50 |
| chr4_-_15003854 | 2.29 |
ENSDART00000134701
ENSDART00000002401 |
klhdc10
|
kelch domain containing 10 |
| chr3_+_34988670 | 2.29 |
ENSDART00000011319
|
smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr5_+_24047292 | 2.28 |
ENSDART00000029889
|
ctdnep1a
|
CTD nuclear envelope phosphatase 1a |
| chr21_-_44731865 | 2.26 |
ENSDART00000013814
|
ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 homolog (human) |
| chr15_-_47107557 | 2.26 |
ENSDART00000111880
|
CABZ01079080.1
|
|
| chr1_+_31573225 | 2.25 |
ENSDART00000075286
|
slc2a15b
|
solute carrier family 2 (facilitated glucose transporter), member 15b |
| chr15_+_23951560 | 2.23 |
ENSDART00000191133
|
myo18ab
|
myosin XVIIIAb |
| chr17_+_43863708 | 2.23 |
ENSDART00000133874
ENSDART00000140316 ENSDART00000142929 ENSDART00000148090 |
zgc:66313
|
zgc:66313 |
| chr1_-_58900851 | 2.22 |
ENSDART00000183085
ENSDART00000188855 ENSDART00000182567 |
CABZ01084501.3
|
Danio rerio microfibril-associated glycoprotein 4-like (LOC100334800), transcript variant 2, mRNA. |
| chr20_+_23501535 | 2.19 |
ENSDART00000177922
ENSDART00000058532 |
palld
|
palladin, cytoskeletal associated protein |
| chr12_-_13729263 | 2.13 |
ENSDART00000078021
|
foxh1
|
forkhead box H1 |
| chr10_+_37182626 | 2.13 |
ENSDART00000137636
|
ksr1a
|
kinase suppressor of ras 1a |
| chr10_-_33251876 | 2.08 |
ENSDART00000184565
|
bcl7ba
|
BCL tumor suppressor 7Ba |
| chr10_-_36793412 | 2.03 |
ENSDART00000185966
|
dhrs13a.2
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 2 |
| chr6_-_46742455 | 2.02 |
ENSDART00000011970
|
zgc:66479
|
zgc:66479 |
| chr5_+_24520437 | 2.01 |
ENSDART00000144560
|
dgcr8
|
DGCR8 microprocessor complex subunit |
| chr18_+_15795748 | 1.98 |
ENSDART00000147024
ENSDART00000137681 |
nudt4a
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4a |
| chr15_-_28908027 | 1.97 |
ENSDART00000182790
ENSDART00000192461 |
eml2
|
echinoderm microtubule associated protein like 2 |
| chr16_+_53278406 | 1.97 |
ENSDART00000010792
|
ptdss1a
|
phosphatidylserine synthase 1a |
| chr20_-_15161669 | 1.97 |
ENSDART00000080333
ENSDART00000063882 |
plpp6
|
phospholipid phosphatase 6 |
| chr16_+_46401756 | 1.96 |
ENSDART00000147370
ENSDART00000144000 |
rpz2
|
rapunzel 2 |
| chr15_-_28907709 | 1.96 |
ENSDART00000017268
|
eml2
|
echinoderm microtubule associated protein like 2 |
| chr23_+_36308428 | 1.95 |
ENSDART00000134607
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr1_+_496268 | 1.92 |
ENSDART00000109415
|
blzf1
|
basic leucine zipper nuclear factor 1 |
| chr9_+_22782027 | 1.92 |
ENSDART00000090816
|
rif1
|
replication timing regulatory factor 1 |
| chr7_-_18881358 | 1.92 |
ENSDART00000021502
|
mllt3
|
MLLT3, super elongation complex subunit |
| chr13_-_25581303 | 1.90 |
ENSDART00000087533
|
csgalnact2
|
chondroitin sulfate N-acetylgalactosaminyltransferase 2 |
| chr14_-_5407555 | 1.89 |
ENSDART00000001424
|
pcgf1
|
polycomb group ring finger 1 |
| chr12_-_26538823 | 1.89 |
ENSDART00000143213
|
acsf2
|
acyl-CoA synthetase family member 2 |
| chr3_+_33745014 | 1.88 |
ENSDART00000159966
|
nacc1a
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing a |
| chr21_-_18275226 | 1.86 |
ENSDART00000126672
ENSDART00000135239 |
brd3a
|
bromodomain containing 3a |
| chr18_+_173603 | 1.86 |
ENSDART00000185918
|
larp6a
|
La ribonucleoprotein domain family, member 6a |
| chr19_-_31765615 | 1.84 |
ENSDART00000103636
|
si:dkeyp-120h9.1
|
si:dkeyp-120h9.1 |
| chr14_+_38786298 | 1.84 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr17_+_50261603 | 1.84 |
ENSDART00000154503
ENSDART00000154467 |
syncripl
|
synaptotagmin binding, cytoplasmic RNA interacting protein, like |
| chr7_+_7027199 | 1.84 |
ENSDART00000193180
|
rbm14b
|
RNA binding motif protein 14b |
| chr7_+_30779761 | 1.83 |
ENSDART00000066806
ENSDART00000173671 |
mcee
|
methylmalonyl CoA epimerase |
| chr17_-_24937879 | 1.82 |
ENSDART00000153964
|
CR391986.1
|
|
| chr18_+_15778110 | 1.78 |
ENSDART00000014188
|
ube2na
|
ubiquitin-conjugating enzyme E2Na |
| chr15_-_28904371 | 1.78 |
ENSDART00000155154
|
eml2
|
echinoderm microtubule associated protein like 2 |
| chr15_-_26089012 | 1.78 |
ENSDART00000152243
ENSDART00000140214 |
dph1
|
diphthamide biosynthesis 1 |
| chr2_-_31833347 | 1.78 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr8_-_39859688 | 1.77 |
ENSDART00000019907
|
unc119.1
|
unc-119 homolog 1 |
| chr6_-_11792152 | 1.77 |
ENSDART00000183403
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr16_+_30002605 | 1.77 |
ENSDART00000160555
|
sema6e
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6E |
| chr6_+_23810529 | 1.76 |
ENSDART00000166921
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr6_+_19950107 | 1.76 |
ENSDART00000181632
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr7_+_32693890 | 1.76 |
ENSDART00000121972
|
slc39a13
|
solute carrier family 39 (zinc transporter), member 13 |
| chr3_-_40051425 | 1.76 |
ENSDART00000146700
|
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr18_+_18000695 | 1.76 |
ENSDART00000146898
|
si:ch211-212o1.2
|
si:ch211-212o1.2 |
| chr14_+_12178915 | 1.76 |
ENSDART00000054626
|
hdac3
|
histone deacetylase 3 |
| chr21_+_25765734 | 1.75 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr2_-_55337585 | 1.75 |
ENSDART00000177924
|
tpm4b
|
tropomyosin 4b |
| chr25_-_7670391 | 1.74 |
ENSDART00000044970
|
bet1l
|
Bet1 golgi vesicular membrane trafficking protein-like |
| chr7_-_41881177 | 1.74 |
ENSDART00000174258
ENSDART00000018972 |
zgc:92818
|
zgc:92818 |
| chr24_+_21346796 | 1.73 |
ENSDART00000126519
|
shisa2b
|
shisa family member 2b |
| chr11_-_25461336 | 1.73 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr1_-_58887610 | 1.73 |
ENSDART00000180647
|
CABZ01084501.1
|
microfibril-associated glycoprotein 4-like precursor |
| chr14_-_5407118 | 1.72 |
ENSDART00000168074
|
pcgf1
|
polycomb group ring finger 1 |
| chr14_-_763744 | 1.71 |
ENSDART00000165856
|
trim35-27
|
tripartite motif containing 35-27 |
| chr8_-_32354677 | 1.71 |
ENSDART00000138268
ENSDART00000133245 ENSDART00000179677 ENSDART00000174450 |
ipo11
|
importin 11 |
| chr24_+_19414561 | 1.71 |
ENSDART00000193157
|
sulf1
|
sulfatase 1 |
| chr8_-_11016766 | 1.70 |
ENSDART00000064066
|
bcas2
|
BCAS2, pre-mRNA processing factor |
| chr24_+_14937205 | 1.70 |
ENSDART00000091735
|
dok6
|
docking protein 6 |
| chr9_+_34397843 | 1.69 |
ENSDART00000146314
|
med14
|
mediator complex subunit 14 |
| chr20_-_15161502 | 1.68 |
ENSDART00000187072
|
plpp6
|
phospholipid phosphatase 6 |
| chr15_-_16946124 | 1.67 |
ENSDART00000154923
|
hip1
|
huntingtin interacting protein 1 |
| chr17_+_45009868 | 1.65 |
ENSDART00000085009
|
coq6
|
coenzyme Q6 monooxygenase |
| chr17_+_6793001 | 1.65 |
ENSDART00000030773
|
foxo3a
|
forkhead box O3A |
| chr9_+_33154841 | 1.64 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr7_-_30553588 | 1.64 |
ENSDART00000139546
|
sltm
|
SAFB-like, transcription modulator |
| chr18_-_15610856 | 1.64 |
ENSDART00000099849
|
arntl2
|
aryl hydrocarbon receptor nuclear translocator-like 2 |
| chr12_-_10508952 | 1.63 |
ENSDART00000152806
|
zgc:152977
|
zgc:152977 |
| chr23_+_384850 | 1.62 |
ENSDART00000114000
|
zgc:101663
|
zgc:101663 |
| chr5_-_13848296 | 1.62 |
ENSDART00000127109
|
figla
|
folliculogenesis specific bHLH transcription factor |
| chr6_-_7686594 | 1.62 |
ENSDART00000091836
ENSDART00000151697 |
ubn2a
|
ubinuclein 2a |
| chr2_-_11027258 | 1.61 |
ENSDART00000081072
ENSDART00000193824 ENSDART00000187036 ENSDART00000097741 |
ssbp3a
|
single stranded DNA binding protein 3a |
| chr23_-_36446307 | 1.61 |
ENSDART00000136623
|
zgc:174906
|
zgc:174906 |
| chr6_+_3717613 | 1.61 |
ENSDART00000184330
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr12_-_41759686 | 1.61 |
ENSDART00000172175
ENSDART00000165152 |
ppp2r2d
|
protein phosphatase 2, regulatory subunit B, delta |
| chr2_-_26596794 | 1.60 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr11_+_31609481 | 1.60 |
ENSDART00000124830
ENSDART00000162768 |
zgc:162816
|
zgc:162816 |
| chr8_-_18613948 | 1.60 |
ENSDART00000089172
|
cpox
|
coproporphyrinogen oxidase |
| chr5_+_36661058 | 1.58 |
ENSDART00000125653
|
capns1a
|
calpain, small subunit 1 a |
| chr23_-_45487304 | 1.58 |
ENSDART00000148889
|
znhit6
|
zinc finger HIT-type containing 6 |
| chr17_+_32622933 | 1.58 |
ENSDART00000077418
|
ctsba
|
cathepsin Ba |
| chr22_-_4769140 | 1.57 |
ENSDART00000165235
|
calr3a
|
calreticulin 3a |
| chr12_-_7234915 | 1.57 |
ENSDART00000048866
|
ipmkb
|
inositol polyphosphate multikinase b |
| chr8_-_38022298 | 1.56 |
ENSDART00000067809
|
rab11fip1a
|
RAB11 family interacting protein 1 (class I) a |
| chr24_+_19415124 | 1.56 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr21_+_37513488 | 1.55 |
ENSDART00000185394
|
amot
|
angiomotin |
| chr15_-_33304133 | 1.54 |
ENSDART00000186092
|
nbeab
|
neurobeachin b |
| chr7_+_44802353 | 1.54 |
ENSDART00000066380
|
ca7
|
carbonic anhydrase VII |
| chr7_+_55950229 | 1.54 |
ENSDART00000082780
|
ACSF3
|
acyl-CoA synthetase family member 3 |
| chr17_+_584369 | 1.54 |
ENSDART00000165143
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr17_+_19626479 | 1.53 |
ENSDART00000044993
ENSDART00000131863 |
rgs7a
|
regulator of G protein signaling 7a |
| chr9_-_23807032 | 1.53 |
ENSDART00000027443
|
esyt3
|
extended synaptotagmin-like protein 3 |
| chr1_-_40519340 | 1.52 |
ENSDART00000114659
|
maml3
|
mastermind-like transcriptional coactivator 3 |
| chr23_+_28381260 | 1.52 |
ENSDART00000162722
|
zgc:153867
|
zgc:153867 |
| chr9_-_16239032 | 1.52 |
ENSDART00000131398
ENSDART00000138204 |
myo1b
|
myosin IB |
| chr7_+_33424044 | 1.51 |
ENSDART00000180260
|
glceb
|
glucuronic acid epimerase b |
| chr15_+_30310843 | 1.51 |
ENSDART00000112784
|
lyrm9
|
LYR motif containing 9 |
| chr5_-_69934558 | 1.51 |
ENSDART00000124954
|
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr18_+_18000887 | 1.51 |
ENSDART00000147797
|
si:ch211-212o1.2
|
si:ch211-212o1.2 |
| chr17_+_52300018 | 1.50 |
ENSDART00000190302
|
esrrb
|
estrogen-related receptor beta |
| chr6_+_1724889 | 1.50 |
ENSDART00000157415
|
acvr2ab
|
activin A receptor type 2Ab |
| chr11_+_13024002 | 1.50 |
ENSDART00000104113
|
btf3l4
|
basic transcription factor 3-like 4 |
| chr19_+_40071561 | 1.50 |
ENSDART00000191109
|
zmym4
|
zinc finger, MYM-type 4 |
| chr11_-_39118882 | 1.49 |
ENSDART00000113185
ENSDART00000156526 |
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr2_-_43583896 | 1.49 |
ENSDART00000161711
|
itgb1b
|
integrin, beta 1b |
| chr6_-_39521832 | 1.48 |
ENSDART00000065038
|
atf1
|
activating transcription factor 1 |
| chr4_+_13586455 | 1.48 |
ENSDART00000187230
|
tnpo3
|
transportin 3 |
| chr8_-_944055 | 1.48 |
ENSDART00000092773
|
mrps27
|
mitochondrial ribosomal protein S27 |
| chr13_+_32454262 | 1.48 |
ENSDART00000057421
|
rdh14a
|
retinol dehydrogenase 14a |
| chr10_+_1052591 | 1.48 |
ENSDART00000123405
|
unc5c
|
unc-5 netrin receptor C |
| chr10_-_44017642 | 1.47 |
ENSDART00000135240
ENSDART00000014669 |
acads
|
acyl-CoA dehydrogenase short chain |
| chr12_-_31724198 | 1.47 |
ENSDART00000153056
ENSDART00000165299 ENSDART00000137464 ENSDART00000080173 |
srsf2a
|
serine/arginine-rich splicing factor 2a |
| chr6_-_33916756 | 1.47 |
ENSDART00000137447
ENSDART00000138488 |
nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr2_-_21170517 | 1.47 |
ENSDART00000135417
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr6_+_52212927 | 1.46 |
ENSDART00000143458
|
ywhaba
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide a |
| chr17_-_8673278 | 1.46 |
ENSDART00000171850
ENSDART00000017337 ENSDART00000148504 ENSDART00000148808 |
ctbp2a
|
C-terminal binding protein 2a |
| chr22_-_7025393 | 1.46 |
ENSDART00000003422
|
smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr3_+_7771420 | 1.46 |
ENSDART00000156809
ENSDART00000156309 |
hook2
|
hook microtubule-tethering protein 2 |
| chr19_+_30867845 | 1.45 |
ENSDART00000047461
|
mfsd2ab
|
major facilitator superfamily domain containing 2ab |
| chr13_+_28618086 | 1.44 |
ENSDART00000087001
|
cnnm2a
|
cyclin and CBS domain divalent metal cation transport mediator 2a |
| chr1_-_9527200 | 1.44 |
ENSDART00000110790
|
si:ch73-12o23.1
|
si:ch73-12o23.1 |
| chr5_-_54712159 | 1.44 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr17_-_43863700 | 1.44 |
ENSDART00000157530
|
ahsa1b
|
AHA1, activator of heat shock protein ATPase homolog 1b |
| chr8_+_17143501 | 1.43 |
ENSDART00000061758
|
mier3b
|
mesoderm induction early response 1, family member 3 b |
| chr12_+_19959037 | 1.43 |
ENSDART00000015780
|
ercc4
|
excision repair cross-complementation group 4 |
| chr10_-_22150419 | 1.43 |
ENSDART00000006173
|
cldn7b
|
claudin 7b |
| chr5_+_54497475 | 1.43 |
ENSDART00000158149
ENSDART00000163968 ENSDART00000160446 |
tmem203
|
transmembrane protein 203 |
| chr23_-_27571667 | 1.43 |
ENSDART00000008174
|
pfkma
|
phosphofructokinase, muscle a |
| chr7_-_69795488 | 1.42 |
ENSDART00000162414
|
USP53 (1 of many)
|
ubiquitin specific peptidase 53 |
| chr17_+_46387086 | 1.42 |
ENSDART00000157079
|
si:dkey-206p8.1
|
si:dkey-206p8.1 |
| chr17_-_32779556 | 1.41 |
ENSDART00000077459
|
smyd2a
|
SET and MYND domain containing 2a |
| chr14_+_22129096 | 1.41 |
ENSDART00000132514
|
ccng1
|
cyclin G1 |
| chr20_-_44576949 | 1.41 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr8_-_49207319 | 1.40 |
ENSDART00000022870
|
fam110a
|
family with sequence similarity 110, member A |
| chr21_+_43702016 | 1.40 |
ENSDART00000017176
|
dkc1
|
dyskeratosis congenita 1, dyskerin |
| chr16_+_27543893 | 1.39 |
ENSDART00000182421
|
si:ch211-197h24.6
|
si:ch211-197h24.6 |
| chr20_-_34801181 | 1.39 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr6_-_39903393 | 1.39 |
ENSDART00000085945
|
tsfm
|
Ts translation elongation factor, mitochondrial |
| chr24_-_9002038 | 1.39 |
ENSDART00000066783
ENSDART00000150185 |
mppe1
|
metallophosphoesterase 1 |
| chr8_-_19467011 | 1.39 |
ENSDART00000162010
|
zgc:92140
|
zgc:92140 |
| chr6_+_59832786 | 1.38 |
ENSDART00000154985
ENSDART00000102148 |
ddx3b
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3b |
| chr3_-_23574622 | 1.37 |
ENSDART00000176012
|
igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr22_-_15437242 | 1.37 |
ENSDART00000063719
|
rpp21
|
ribonuclease P 21 subunit |
| chr24_+_36018164 | 1.36 |
ENSDART00000182815
ENSDART00000126941 |
gnal2
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type 2 |
| chr22_-_20419660 | 1.36 |
ENSDART00000105520
|
pias4a
|
protein inhibitor of activated STAT, 4a |
| chr23_+_20431388 | 1.36 |
ENSDART00000132920
ENSDART00000102963 ENSDART00000109899 ENSDART00000140219 |
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr10_+_1849874 | 1.35 |
ENSDART00000158897
ENSDART00000149956 |
apc
|
adenomatous polyposis coli |
| chr24_+_9298198 | 1.35 |
ENSDART00000165780
|
otud1
|
OTU deubiquitinase 1 |
| chr23_+_25291891 | 1.35 |
ENSDART00000016248
|
pa2g4b
|
proliferation-associated 2G4, b |
| chr7_-_18877109 | 1.34 |
ENSDART00000113593
|
mllt3
|
MLLT3, super elongation complex subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of yy1a+yy1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.7 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.9 | 2.7 | GO:0034770 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.8 | 3.3 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.7 | 2.1 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.7 | 2.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.7 | 2.0 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.6 | 3.0 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.6 | 3.5 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.5 | 3.3 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.5 | 2.7 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.5 | 1.6 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.5 | 4.9 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.5 | 1.4 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.5 | 1.4 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.5 | 2.8 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.5 | 2.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.4 | 2.1 | GO:1901908 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.4 | 2.5 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.4 | 3.3 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.4 | 1.2 | GO:0033512 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.4 | 1.2 | GO:1901255 | nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.4 | 2.0 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.4 | 3.6 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.4 | 1.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.4 | 2.3 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 0.4 | 1.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.4 | 1.8 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) regulation of cellular amine metabolic process(GO:0033238) |
| 0.4 | 1.5 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.4 | 1.5 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.4 | 5.0 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.4 | 2.8 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.3 | 1.0 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.3 | 1.0 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.3 | 1.0 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.3 | 2.0 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.3 | 2.3 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.3 | 1.6 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.3 | 0.6 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.3 | 2.3 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.3 | 1.6 | GO:0046416 | D-amino acid catabolic process(GO:0019478) D-serine catabolic process(GO:0036088) D-amino acid metabolic process(GO:0046416) D-serine metabolic process(GO:0070178) |
| 0.3 | 1.3 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.3 | 0.9 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.3 | 2.7 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.3 | 0.9 | GO:0019408 | dolichol biosynthetic process(GO:0019408) |
| 0.3 | 0.9 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.3 | 1.8 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.3 | 1.8 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.3 | 0.3 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.3 | 1.4 | GO:0045056 | transcytosis(GO:0045056) |
| 0.3 | 1.1 | GO:0072149 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.3 | 2.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.3 | 1.1 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.3 | 0.8 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.3 | 1.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.3 | 1.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.3 | 2.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.3 | 1.4 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.3 | 1.4 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.3 | 0.8 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.3 | 0.8 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.3 | 1.6 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.3 | 1.3 | GO:0039694 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.3 | 1.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.3 | 1.0 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.3 | 2.8 | GO:0045453 | bone resorption(GO:0045453) |
| 0.3 | 1.0 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.3 | 1.8 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.2 | 0.7 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.2 | 0.5 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.2 | 1.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.2 | 0.7 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.2 | 1.0 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.2 | 1.9 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.2 | 2.6 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.2 | 0.7 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.2 | 0.9 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.2 | 1.8 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.2 | 0.9 | GO:2000252 | negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.2 | 0.7 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.2 | 2.4 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.2 | 1.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.2 | 1.9 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.2 | 1.9 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.2 | 2.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 2.3 | GO:0006735 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 1.5 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.2 | 0.8 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.2 | 0.6 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.2 | 0.8 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.2 | 2.2 | GO:0021707 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 0.4 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.2 | 2.2 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.2 | 0.6 | GO:0006178 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.2 | 0.6 | GO:0032196 | transposition(GO:0032196) |
| 0.2 | 2.2 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.2 | 2.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.2 | 1.0 | GO:0051204 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.2 | 0.6 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.2 | 1.1 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.2 | 0.8 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.2 | 0.6 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 0.6 | GO:1901546 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.2 | 1.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.2 | 0.9 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.2 | 0.7 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.2 | 0.5 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.2 | 0.9 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.2 | 0.9 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.2 | 1.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 | 0.7 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.2 | 1.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 0.7 | GO:0061015 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.2 | 0.2 | GO:0001502 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.2 | 3.4 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 6.7 | GO:0060415 | muscle tissue morphogenesis(GO:0060415) |
| 0.2 | 1.8 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.2 | 0.5 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.2 | 0.8 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.2 | 0.5 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.2 | 0.5 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.2 | 1.6 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 1.4 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.2 | 1.4 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.2 | 0.6 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 0.5 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.2 | 0.3 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.2 | 1.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.2 | 2.6 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 0.9 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.2 | 0.8 | GO:0006901 | vesicle coating(GO:0006901) vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.2 | 0.5 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.2 | 0.6 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.2 | 1.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.2 | 1.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 0.9 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 | 4.2 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 1.2 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.1 | 1.2 | GO:0016246 | RNA interference(GO:0016246) |
| 0.1 | 1.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 2.6 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 1.5 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 6.5 | GO:0071166 | ribonucleoprotein complex localization(GO:0071166) |
| 0.1 | 1.7 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 1.2 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.4 | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.1 | 1.3 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 2.1 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 1.4 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.1 | 0.7 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.1 | 0.5 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.1 | 8.5 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.1 | 0.6 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 1.6 | GO:0008585 | female gonad development(GO:0008585) |
| 0.1 | 0.7 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.5 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) |
| 0.1 | 0.6 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.1 | 1.6 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 | 0.6 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 1.0 | GO:0014028 | notochord formation(GO:0014028) |
| 0.1 | 0.6 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 3.6 | GO:0098727 | maintenance of cell number(GO:0098727) |
| 0.1 | 0.8 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.6 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 0.6 | GO:0072575 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.1 | 0.9 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.5 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 0.8 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.6 | GO:0019627 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) ammonia homeostasis(GO:0097272) |
| 0.1 | 1.5 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 1.4 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 | 1.0 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 2.0 | GO:0016233 | telomere capping(GO:0016233) |
| 0.1 | 0.9 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.1 | 0.3 | GO:0042823 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 | 1.0 | GO:2000758 | positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.1 | 1.8 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.3 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 0.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.3 | GO:0046831 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.1 | 0.2 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.1 | 0.4 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 0.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 1.9 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.5 | GO:0010991 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 1.0 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.1 | 0.6 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.1 | 0.3 | GO:0043525 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.1 | 1.2 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 | 1.1 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 1.3 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 4.4 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 0.3 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.1 | 1.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.9 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 1.2 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.1 | 0.5 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 1.1 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.8 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.1 | 0.3 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 1.9 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.1 | 1.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 2.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 1.8 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 0.7 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 0.3 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 2.5 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.1 | 1.6 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.6 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.4 | GO:0030329 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.1 | 2.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 0.5 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 9.5 | GO:0048545 | response to steroid hormone(GO:0048545) |
| 0.1 | 1.6 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.1 | 0.3 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 1.8 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.6 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 0.2 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.1 | 0.9 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 1.8 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 2.9 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 1.7 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 1.0 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 2.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.6 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 2.9 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 0.6 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 1.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.3 | GO:0090387 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.1 | 0.5 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.1 | 0.2 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.1 | 0.4 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 1.0 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 0.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 1.7 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.1 | 0.3 | GO:0032615 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.1 | 0.4 | GO:0034501 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.1 | 0.5 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 1.0 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 0.4 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.1 | 0.2 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.1 | 0.3 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 0.6 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.8 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.1 | 2.6 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.1 | 2.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 0.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.3 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 1.6 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.1 | 1.4 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 1.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.6 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 | 1.1 | GO:0030330 | DNA damage response, signal transduction by p53 class mediator(GO:0030330) |
| 0.1 | 0.3 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 1.7 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.1 | 0.1 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 0.4 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 2.2 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.1 | 1.0 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 0.7 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 0.9 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 3.1 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.1 | 1.0 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 2.3 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.1 | 1.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.4 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.1 | 0.4 | GO:0048662 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.1 | 0.6 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.4 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 0.5 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.2 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 1.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 1.0 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 0.3 | GO:1904398 | regulation of neuromuscular junction development(GO:1904396) positive regulation of neuromuscular junction development(GO:1904398) |
| 0.1 | 0.6 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.1 | 3.1 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.6 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.1 | 0.8 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.2 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 0.2 | GO:0051444 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.1 | 0.7 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.1 | 0.2 | GO:0060043 | regulation of cardiac muscle tissue growth(GO:0055021) regulation of cardiac muscle cell proliferation(GO:0060043) regulation of heart growth(GO:0060420) |
| 0.1 | 0.4 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 0.4 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 1.7 | GO:0051236 | nucleic acid transport(GO:0050657) RNA transport(GO:0050658) establishment of RNA localization(GO:0051236) |
| 0.1 | 0.4 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 0.6 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 1.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.8 | GO:0060021 | palate development(GO:0060021) |
| 0.1 | 0.2 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.4 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.0 | 0.7 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.0 | GO:0060827 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.0 | 7.0 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.1 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.0 | 0.4 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 1.6 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 0.7 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 1.6 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.6 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 1.9 | GO:0051057 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 0.1 | GO:0016441 | posttranscriptional gene silencing(GO:0016441) posttranscriptional gene silencing by RNA(GO:0035194) |
| 0.0 | 1.2 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.6 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 4.1 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.1 | GO:0010359 | regulation of anion channel activity(GO:0010359) |
| 0.0 | 0.1 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 5.4 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.4 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 1.3 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 1.4 | GO:0032200 | telomere maintenance(GO:0000723) telomere organization(GO:0032200) |
| 0.0 | 0.6 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.0 | 1.1 | GO:0050433 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) catecholamine secretion(GO:0050432) regulation of catecholamine secretion(GO:0050433) |
| 0.0 | 2.1 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.3 | GO:0002295 | T-helper cell lineage commitment(GO:0002295) T cell lineage commitment(GO:0002360) alpha-beta T cell lineage commitment(GO:0002363) CD4-positive or CD8-positive, alpha-beta T cell lineage commitment(GO:0043369) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) |
| 0.0 | 3.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.3 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 1.4 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 1.0 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 1.2 | GO:0006101 | citrate metabolic process(GO:0006101) tricarboxylic acid metabolic process(GO:0072350) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.4 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.9 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 1.7 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 1.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0060465 | pharynx development(GO:0060465) |
| 0.0 | 0.4 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.4 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 0.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.8 | GO:0009081 | branched-chain amino acid metabolic process(GO:0009081) |
| 0.0 | 0.1 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.2 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.0 | 1.1 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 3.2 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.1 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.3 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.7 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 1.2 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.8 | GO:1902749 | regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.0 | 0.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.6 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.4 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.6 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 0.1 | GO:0002691 | regulation of cellular extravasation(GO:0002691) positive regulation of cellular extravasation(GO:0002693) positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 1.8 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.1 | GO:0032602 | chemokine production(GO:0032602) thymocyte migration(GO:0072679) |
| 0.0 | 0.3 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.1 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.0 | 7.3 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 0.5 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) neutral lipid catabolic process(GO:0046461) acylglycerol catabolic process(GO:0046464) |
| 0.0 | 0.2 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 | 0.1 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.2 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.1 | GO:0031060 | regulation of histone methylation(GO:0031060) |
| 0.0 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.6 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 1.1 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.6 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.2 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.0 | 0.4 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.7 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.0 | 0.5 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.3 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.0 | 0.1 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.4 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.2 | GO:0014829 | vascular smooth muscle contraction(GO:0014829) |
| 0.0 | 1.2 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 1.5 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.1 | GO:0055014 | atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.4 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.1 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.8 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.3 | GO:1901796 | regulation of signal transduction by p53 class mediator(GO:1901796) |
| 0.0 | 1.1 | GO:0006354 | DNA-templated transcription, elongation(GO:0006354) |
| 0.0 | 0.1 | GO:0016571 | histone methylation(GO:0016571) |
| 0.0 | 0.2 | GO:0090224 | regulation of spindle organization(GO:0090224) |
| 0.0 | 1.1 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.1 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.1 | GO:0098868 | endochondral bone growth(GO:0003416) growth plate cartilage development(GO:0003417) bone growth(GO:0098868) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.9 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.2 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.8 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.1 | GO:1902260 | negative regulation of potassium ion transport(GO:0043267) negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.7 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.7 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.3 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.2 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.8 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.5 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.1 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.2 | GO:0031274 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.7 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 1.1 | GO:0006417 | regulation of translation(GO:0006417) |
| 0.0 | 0.3 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 1.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.1 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 3.9 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.2 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.4 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.6 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0071715 | icosanoid transport(GO:0071715) fatty acid derivative transport(GO:1901571) |
| 0.0 | 0.7 | GO:0007088 | regulation of mitotic nuclear division(GO:0007088) |
| 0.0 | 0.1 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.3 | GO:0051169 | nucleocytoplasmic transport(GO:0006913) nuclear transport(GO:0051169) |
| 0.0 | 0.8 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.3 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.2 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.0 | 0.1 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 0.0 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.5 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 0.2 | GO:0071453 | cellular response to decreased oxygen levels(GO:0036294) cellular response to oxygen levels(GO:0071453) cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.3 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.2 | GO:0000272 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.6 | GO:0005977 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.0 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) negative regulation of anion transport(GO:1903792) |
| 0.0 | 0.3 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.1 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.1 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.4 | GO:0016311 | dephosphorylation(GO:0016311) |
| 0.0 | 0.1 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.0 | 0.3 | GO:0046427 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.0 | 0.1 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 0.2 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.1 | GO:0071407 | cellular response to organic cyclic compound(GO:0071407) |
| 0.0 | 0.2 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.6 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.1 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.1 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.4 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.4 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.4 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.2 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.1 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.0 | 1.7 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.0 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.9 | 0.9 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.6 | 3.0 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.5 | 3.6 | GO:0030681 | multimeric ribonuclease P complex(GO:0030681) |
| 0.5 | 1.4 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.5 | 1.8 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.4 | 3.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.4 | 1.2 | GO:1990879 | CST complex(GO:1990879) |
| 0.4 | 3.7 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.4 | 2.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.4 | 2.5 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.4 | 1.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.3 | 1.4 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.3 | 1.4 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.3 | 1.6 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.3 | 3.0 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.3 | 2.6 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.3 | 1.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.3 | 1.3 | GO:0034657 | GID complex(GO:0034657) |
| 0.2 | 1.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 1.4 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.2 | 0.7 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.2 | 5.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 0.9 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.2 | 2.8 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.2 | 2.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 0.8 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.2 | 1.4 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 1.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.2 | 0.8 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.2 | 3.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 7.2 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 0.9 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.2 | 0.9 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.2 | 0.7 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.2 | 1.4 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.2 | 0.7 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.2 | 0.5 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.2 | 0.5 | GO:0019185 | snRNA-activating protein complex(GO:0019185) |
| 0.2 | 0.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.2 | 2.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 2.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.4 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.9 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.6 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 1.1 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 1.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.7 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 0.8 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 9.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 2.0 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 1.3 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 1.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 4.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 2.6 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.1 | 1.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.7 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 1.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.7 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.1 | 0.9 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 1.3 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.4 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 0.7 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.1 | 1.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 2.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 0.5 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 2.0 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.6 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.4 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 2.5 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 1.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.6 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.1 | 0.3 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 3.4 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.1 | 1.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.3 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.1 | 0.5 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 11.5 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 1.7 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 0.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 5.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.6 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 0.5 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 1.0 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.5 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.1 | 2.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 0.3 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.9 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 1.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 1.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.7 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 1.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.7 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 2.4 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 1.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 2.8 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 1.6 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.1 | 0.8 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.7 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 0.3 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 4.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.2 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 0.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 4.0 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 1.1 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 3.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.2 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.2 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 4.3 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.9 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.6 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 1.0 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.8 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 2.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.3 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 1.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 2.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 2.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.6 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 1.0 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 2.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 3.7 | GO:0098791 | Golgi subcompartment(GO:0098791) |
| 0.0 | 0.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.8 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.6 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0098753 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) anchored component of the cytoplasmic side of the plasma membrane(GO:0098753) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.3 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.4 | GO:0070461 | SAGA-type complex(GO:0070461) |
| 0.0 | 1.0 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.1 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.0 | 0.4 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 1.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 2.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.3 | GO:0030120 | vesicle coat(GO:0030120) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.1 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 0.2 | GO:0000798 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 1.3 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.0 | 1.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.1 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.8 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 1.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.7 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 2.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 9.3 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 2.1 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.3 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.9 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 0.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.3 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.5 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 2.3 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 1.5 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 7.9 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.6 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.9 | 2.7 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.7 | 2.7 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.6 | 1.9 | GO:0031956 | medium-chain fatty acid-CoA ligase activity(GO:0031956) butyrate-CoA ligase activity(GO:0047760) |
| 0.5 | 2.2 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.5 | 1.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.5 | 1.6 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.5 | 4.0 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.5 | 4.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.4 | 2.1 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.4 | 2.9 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.4 | 1.9 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.4 | 1.5 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.4 | 1.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.4 | 1.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 1.7 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.3 | 2.4 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.3 | 1.4 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.3 | 1.6 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.3 | 1.6 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.3 | 0.9 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.3 | 1.5 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.3 | 0.9 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.3 | 1.5 | GO:0098634 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.3 | 0.9 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.3 | 1.8 | GO:0097363 | protein N-acetylglucosaminyltransferase activity(GO:0016262) protein O-GlcNAc transferase activity(GO:0097363) |
| 0.3 | 1.1 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.3 | 1.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.3 | 0.8 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.3 | 1.1 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.3 | 1.8 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.3 | 0.8 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.3 | 1.0 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.3 | 1.8 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 0.7 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.2 | 1.7 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.2 | 2.6 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.2 | 0.7 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.2 | 1.4 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.2 | 0.7 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.2 | 0.7 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.2 | 0.9 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 1.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.2 | 5.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 0.9 | GO:0045547 | dehydrodolichyl diphosphate synthase activity(GO:0045547) |
| 0.2 | 1.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 2.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 3.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 0.8 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.2 | 2.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 0.8 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.2 | 1.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 0.6 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.2 | 3.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.2 | 0.6 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.2 | 3.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 1.6 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 0.6 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.2 | 0.8 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.2 | 0.6 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) |
| 0.2 | 0.6 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.2 | 1.8 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.2 | 1.8 | GO:0004549 | tRNA-specific ribonuclease activity(GO:0004549) |
| 0.2 | 2.9 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.2 | 0.8 | GO:0008311 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.2 | 0.5 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.2 | 1.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 0.6 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.2 | 2.7 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.2 | 2.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 0.5 | GO:0001047 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.2 | 2.5 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.2 | 0.8 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.6 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.6 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 3.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 5.3 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.1 | 1.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.9 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.8 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 0.7 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.1 | 0.4 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 2.9 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 0.6 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 1.5 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 2.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 2.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 5.2 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 0.6 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.5 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.6 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 1.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.7 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 3.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.9 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 1.2 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 2.2 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 1.0 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.0 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 0.8 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.3 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 0.1 | 6.9 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 0.5 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.1 | 1.2 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.1 | 1.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 1.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 1.0 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 1.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.4 | GO:0070883 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) pre-miRNA binding(GO:0070883) |
| 0.1 | 0.9 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 2.0 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.4 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 1.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.7 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 1.8 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 0.8 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 1.3 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 1.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 1.7 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 1.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 3.3 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.1 | 1.2 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.1 | 0.9 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 2.3 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 1.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.4 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.1 | 1.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 1.0 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 6.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.3 | GO:0030586 | [methionine synthase] reductase activity(GO:0030586) |
| 0.1 | 1.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 7.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 0.7 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 1.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 6.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 0.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.8 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.2 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.1 | 0.6 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.3 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.1 | 1.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.5 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 1.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 3.0 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 0.4 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 2.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.6 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 0.5 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.8 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 0.6 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 1.1 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.3 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 3.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.5 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.9 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 1.3 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 2.9 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.1 | 0.5 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.3 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.1 | 0.3 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.3 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.2 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.1 | 0.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.2 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.4 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 0.3 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 0.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 2.6 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.1 | 0.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.9 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.2 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.1 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.4 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 0.4 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.2 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 3.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.5 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 1.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 8.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0004555 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 1.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.9 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.6 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.4 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 1.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0031530 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.0 | 1.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.3 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 2.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 2.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 1.1 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.4 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.0 | 1.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.4 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.6 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 2.6 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.2 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 1.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.5 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 4.2 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.4 | GO:0022884 | macromolecule transmembrane transporter activity(GO:0022884) |
| 0.0 | 0.4 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 7.8 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 1.0 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.2 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.4 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 9.0 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 0.4 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 1.4 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 0.9 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.3 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.3 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 1.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 7.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0019767 | immunoglobulin receptor activity(GO:0019763) IgE receptor activity(GO:0019767) |
| 0.0 | 0.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 1.6 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 2.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.6 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 0.3 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.4 | GO:0052744 | phosphatidylinositol-3-phosphatase activity(GO:0004438) phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.0 | 0.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.5 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 0.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 3.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.0 | GO:0031781 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.0 | 7.5 | GO:0003723 | RNA binding(GO:0003723) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.2 | 1.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 3.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 5.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.2 | 3.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 2.9 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 6.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 1.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 2.0 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 2.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 2.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 1.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 1.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 2.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 0.6 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 2.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 2.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 4.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 1.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 0.8 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 0.8 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 0.3 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 1.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 0.7 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 8.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 0.3 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 0.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 1.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 0.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 1.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 1.0 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 1.0 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 0.4 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 1.1 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.6 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 2.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.7 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.5 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.4 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.4 | 4.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.4 | 2.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.4 | 3.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.3 | 4.4 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.2 | 2.5 | REACTOME SIGNALING BY NOTCH | Genes involved in Signaling by NOTCH |
| 0.2 | 2.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.2 | 3.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.2 | 3.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 3.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 4.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 2.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.2 | 1.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 1.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 1.6 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 1.0 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 3.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.2 | 2.8 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.2 | 0.6 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.2 | 0.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 0.5 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 2.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 3.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 2.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 2.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 1.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 1.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 2.8 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 0.4 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 0.9 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 0.8 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 2.9 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 1.4 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 1.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.0 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 1.8 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.6 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 1.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.3 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 1.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.2 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.1 | 0.7 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 0.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.9 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 0.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 3.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 0.3 | REACTOME SIGNALING BY FGFR1 MUTANTS | Genes involved in Signaling by FGFR1 mutants |
| 0.1 | 0.7 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.1 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 0.6 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 1.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.0 | REACTOME NEGATIVE REGULATION OF FGFR SIGNALING | Genes involved in Negative regulation of FGFR signaling |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.9 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.9 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.4 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.4 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.8 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 1.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 4.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.7 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 1.0 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.1 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.9 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.5 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.4 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |