Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
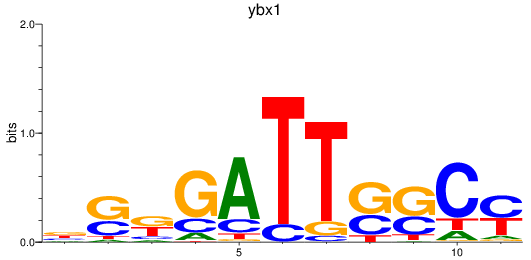
Results for ybx1
Z-value: 0.32
Transcription factors associated with ybx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ybx1
|
ENSDARG00000004757 | Y box binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ybx1 | dr11_v1_chr8_-_47152001_47152086 | 0.81 | 4.4e-05 | Click! |
Activity profile of ybx1 motif
Sorted Z-values of ybx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_6795531 | 0.60 |
ENSDART00000092131
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr15_+_1148074 | 0.54 |
ENSDART00000152638
ENSDART00000152466 ENSDART00000188011 |
mlf1
|
myeloid leukemia factor 1 |
| chr11_-_30352333 | 0.50 |
ENSDART00000030794
|
tmem169a
|
transmembrane protein 169a |
| chr4_-_2545310 | 0.47 |
ENSDART00000150619
ENSDART00000140760 |
e2f7
|
E2F transcription factor 7 |
| chr3_+_62317990 | 0.46 |
ENSDART00000058719
|
znf1124
|
zinc finger protein 1124 |
| chr10_+_36026576 | 0.45 |
ENSDART00000193786
|
hmgb1a
|
high mobility group box 1a |
| chr23_+_6795709 | 0.43 |
ENSDART00000149136
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr10_+_39084354 | 0.42 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr22_+_17248145 | 0.39 |
ENSDART00000136908
|
axdnd1
|
axonemal dynein light chain domain containing 1 |
| chr15_-_19677511 | 0.37 |
ENSDART00000043743
|
si:dkey-4p15.3
|
si:dkey-4p15.3 |
| chr24_+_24923166 | 0.36 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr16_+_4838808 | 0.36 |
ENSDART00000179363
|
ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr21_+_244503 | 0.35 |
ENSDART00000162889
|
stard4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr20_+_16743056 | 0.35 |
ENSDART00000050308
|
calm1b
|
calmodulin 1b |
| chr1_+_50921266 | 0.33 |
ENSDART00000006538
|
otx2a
|
orthodenticle homeobox 2a |
| chr7_+_24391129 | 0.29 |
ENSDART00000108753
|
poln
|
polymerase (DNA directed) nu |
| chr7_-_22981796 | 0.28 |
ENSDART00000167565
|
si:dkey-171c9.3
|
si:dkey-171c9.3 |
| chr14_-_12071679 | 0.27 |
ENSDART00000165581
|
tmsb1
|
thymosin beta 1 |
| chr6_-_34958274 | 0.26 |
ENSDART00000113097
|
hsd17b7
|
hydroxysteroid (17-beta) dehydrogenase 7 |
| chr2_+_33747509 | 0.24 |
ENSDART00000134187
|
si:dkey-31m5.5
|
si:dkey-31m5.5 |
| chr11_-_18604542 | 0.24 |
ENSDART00000171183
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr23_+_44634187 | 0.24 |
ENSDART00000143688
|
si:ch73-265d7.2
|
si:ch73-265d7.2 |
| chr23_-_38054 | 0.22 |
ENSDART00000170393
|
CABZ01074076.1
|
|
| chr2_+_24352497 | 0.22 |
ENSDART00000134909
|
pimr68
|
Pim proto-oncogene, serine/threonine kinase, related 68 |
| chr10_+_43994471 | 0.21 |
ENSDART00000138242
ENSDART00000186359 |
cldn5b
|
claudin 5b |
| chr1_-_20928772 | 0.21 |
ENSDART00000078277
|
msmo1
|
methylsterol monooxygenase 1 |
| chr18_+_44795711 | 0.21 |
ENSDART00000110229
ENSDART00000188262 ENSDART00000139526 |
fam118b
|
family with sequence similarity 118, member B |
| chr19_+_32166702 | 0.20 |
ENSDART00000021798
|
fabp11a
|
fatty acid binding protein 11a |
| chr18_+_21273749 | 0.20 |
ENSDART00000143265
|
hydin
|
HYDIN, axonemal central pair apparatus protein |
| chr11_-_18604317 | 0.20 |
ENSDART00000182081
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr14_+_3038473 | 0.20 |
ENSDART00000026021
ENSDART00000150000 |
cd74a
|
CD74 molecule, major histocompatibility complex, class II invariant chain a |
| chr4_-_16545085 | 0.20 |
ENSDART00000033188
|
btg1
|
B-cell translocation gene 1, anti-proliferative |
| chr25_-_3647277 | 0.20 |
ENSDART00000166363
|
si:ch211-272n13.3
|
si:ch211-272n13.3 |
| chr1_+_26071869 | 0.20 |
ENSDART00000059264
|
mxd4
|
MAX dimerization protein 4 |
| chr9_+_2499627 | 0.19 |
ENSDART00000160782
|
wipf1a
|
WAS/WASL interacting protein family, member 1a |
| chr22_+_38173960 | 0.19 |
ENSDART00000010537
|
cp
|
ceruloplasmin |
| chr5_+_70155935 | 0.19 |
ENSDART00000165570
|
rgs3a
|
regulator of G protein signaling 3a |
| chr1_+_51386649 | 0.18 |
ENSDART00000152289
|
atg4da
|
autophagy related 4D, cysteine peptidase a |
| chr10_+_9159279 | 0.18 |
ENSDART00000064968
|
rasgef1bb
|
RasGEF domain family, member 1Bb |
| chr19_+_770300 | 0.18 |
ENSDART00000062518
|
gstr
|
glutathione S-transferase rho |
| chr12_+_46960579 | 0.18 |
ENSDART00000149032
|
oat
|
ornithine aminotransferase |
| chr25_+_36405021 | 0.18 |
ENSDART00000152801
|
ankrd27
|
ankyrin repeat domain 27 (VPS9 domain) |
| chr24_-_41220538 | 0.18 |
ENSDART00000150207
|
acvr2ba
|
activin A receptor type 2Ba |
| chr3_-_8765165 | 0.18 |
ENSDART00000191131
|
CABZ01058333.1
|
|
| chr25_+_10458990 | 0.18 |
ENSDART00000130354
ENSDART00000044738 |
ric8a
|
RIC8 guanine nucleotide exchange factor A |
| chr10_+_35257651 | 0.18 |
ENSDART00000028940
|
styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr6_-_35446110 | 0.18 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr18_+_50089000 | 0.18 |
ENSDART00000058936
|
scamp5b
|
secretory carrier membrane protein 5b |
| chr5_-_68074592 | 0.17 |
ENSDART00000165052
ENSDART00000018792 |
spag7
|
sperm associated antigen 7 |
| chr9_+_13120419 | 0.17 |
ENSDART00000141005
|
fam117bb
|
family with sequence similarity 117, member Bb |
| chr19_+_56351 | 0.17 |
ENSDART00000168334
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr15_+_43166511 | 0.17 |
ENSDART00000011737
|
flj13639
|
flj13639 |
| chr19_-_1855368 | 0.17 |
ENSDART00000029646
|
rplp1
|
ribosomal protein, large, P1 |
| chr22_-_36875264 | 0.16 |
ENSDART00000137548
|
kng1
|
kininogen 1 |
| chr24_-_39772045 | 0.16 |
ENSDART00000087441
|
GFOD1
|
si:ch211-276f18.2 |
| chr18_+_62932 | 0.16 |
ENSDART00000052638
|
slc27a2a
|
solute carrier family 27 (fatty acid transporter), member 2a |
| chr8_+_53269657 | 0.16 |
ENSDART00000184212
|
gnb1a
|
guanine nucleotide binding protein (G protein), beta polypeptide 1a |
| chr12_+_8822717 | 0.16 |
ENSDART00000021628
|
reep3b
|
receptor accessory protein 3b |
| chr12_-_35885349 | 0.16 |
ENSDART00000085662
|
cep131
|
centrosomal protein 131 |
| chr12_-_33582382 | 0.15 |
ENSDART00000009794
ENSDART00000136617 |
tdrkh
|
tudor and KH domain containing |
| chr7_+_1521834 | 0.15 |
ENSDART00000174007
|
si:cabz01102082.1
|
si:cabz01102082.1 |
| chr25_+_4541211 | 0.15 |
ENSDART00000129978
|
pnpla2
|
patatin-like phospholipase domain containing 2 |
| chr4_+_11375894 | 0.15 |
ENSDART00000190471
ENSDART00000143963 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr18_+_30567945 | 0.15 |
ENSDART00000078894
|
irf8
|
interferon regulatory factor 8 |
| chr23_-_35483163 | 0.14 |
ENSDART00000138660
ENSDART00000113643 ENSDART00000189269 |
fbxo25
|
F-box protein 25 |
| chr4_-_8060962 | 0.14 |
ENSDART00000146622
|
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr4_+_4509996 | 0.14 |
ENSDART00000028694
|
gnsa
|
glucosamine (N-acetyl)-6-sulfatase a |
| chr7_+_22981441 | 0.14 |
ENSDART00000182887
|
ccnb3
|
cyclin B3 |
| chr20_-_54259780 | 0.14 |
ENSDART00000172631
|
fkbp3
|
FK506 binding protein 3 |
| chr1_+_49651016 | 0.14 |
ENSDART00000074380
ENSDART00000101017 |
tsga10
|
testis specific, 10 |
| chr1_+_58840889 | 0.14 |
ENSDART00000098308
|
tmed1b
|
transmembrane p24 trafficking protein 1b |
| chr5_+_31983098 | 0.14 |
ENSDART00000007458
|
ppp6c
|
protein phosphatase 6, catalytic subunit |
| chr25_-_4148719 | 0.14 |
ENSDART00000112880
ENSDART00000023278 |
fads2
|
fatty acid desaturase 2 |
| chr6_+_52350443 | 0.14 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr1_+_54037077 | 0.13 |
ENSDART00000109386
|
triobpa
|
TRIO and F-actin binding protein a |
| chr19_-_6134802 | 0.13 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr14_+_80685 | 0.13 |
ENSDART00000188443
|
stag3
|
stromal antigen 3 |
| chr24_+_7631797 | 0.13 |
ENSDART00000187464
|
cavin1b
|
caveolae associated protein 1b |
| chr7_+_22981909 | 0.13 |
ENSDART00000122449
|
ccnb3
|
cyclin B3 |
| chr3_+_62217479 | 0.13 |
ENSDART00000175012
|
BX470259.4
|
|
| chr20_+_37866171 | 0.13 |
ENSDART00000153190
|
vash2
|
vasohibin 2 |
| chr19_-_26769867 | 0.13 |
ENSDART00000043776
ENSDART00000159489 ENSDART00000138675 |
prrc2a
|
proline-rich coiled-coil 2A |
| chr20_+_47434709 | 0.13 |
ENSDART00000067776
|
rab10
|
RAB10, member RAS oncogene family |
| chr19_-_26770083 | 0.12 |
ENSDART00000193811
ENSDART00000174455 |
prrc2a
|
proline-rich coiled-coil 2A |
| chr25_-_35110695 | 0.12 |
ENSDART00000099859
|
hist1h2a10
|
histone cluster 1 H2A family member 10 |
| chr21_+_19070921 | 0.12 |
ENSDART00000029874
|
nkx6.1
|
NK6 homeobox 1 |
| chr18_+_22606259 | 0.12 |
ENSDART00000128965
|
bcar1
|
breast cancer anti-estrogen resistance 1 |
| chr25_-_3470910 | 0.11 |
ENSDART00000029067
ENSDART00000186737 |
hbp1
|
HMG-box transcription factor 1 |
| chr9_+_29548195 | 0.11 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr23_-_46034609 | 0.11 |
ENSDART00000158712
|
zgc:65873
|
zgc:65873 |
| chr6_-_16981321 | 0.11 |
ENSDART00000154585
|
pimr29
|
Pim proto-oncogene, serine/threonine kinase, related 29 |
| chr4_-_76370630 | 0.11 |
ENSDART00000168831
ENSDART00000174313 |
si:ch73-158p21.3
|
si:ch73-158p21.3 |
| chr22_-_16180467 | 0.11 |
ENSDART00000171331
ENSDART00000185607 |
vcam1b
|
vascular cell adhesion molecule 1b |
| chr1_+_25801648 | 0.10 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr21_-_26490186 | 0.10 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr12_+_10706772 | 0.10 |
ENSDART00000158227
|
top2a
|
DNA topoisomerase II alpha |
| chr24_-_24271629 | 0.10 |
ENSDART00000135060
|
rps6ka3b
|
ribosomal protein S6 kinase, polypeptide 3b |
| chr1_-_50527964 | 0.10 |
ENSDART00000024984
|
papss1
|
3'-phosphoadenosine 5'-phosphosulfate synthase 1 |
| chr7_+_22982201 | 0.10 |
ENSDART00000134116
|
ccnb3
|
cyclin B3 |
| chr21_-_293146 | 0.10 |
ENSDART00000157781
|
plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr17_-_43287290 | 0.10 |
ENSDART00000156885
|
EML5
|
si:dkey-1f12.3 |
| chr3_-_3328097 | 0.10 |
ENSDART00000193140
|
tmem184bb
|
transmembrane protein 184bb |
| chr11_+_35050253 | 0.10 |
ENSDART00000124800
|
fam212aa
|
family with sequence similarity 212, member Aa |
| chr10_+_29816681 | 0.10 |
ENSDART00000100022
|
h2afx1
|
H2A histone family member X1 |
| chr22_-_22301672 | 0.09 |
ENSDART00000111711
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr6_+_16804761 | 0.09 |
ENSDART00000155204
|
pimr6
|
Pim proto-oncogene, serine/threonine kinase, related 6 |
| chr5_+_45138934 | 0.09 |
ENSDART00000041412
ENSDART00000136002 |
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr3_+_62353650 | 0.09 |
ENSDART00000112428
|
iqck
|
IQ motif containing K |
| chr20_-_53366137 | 0.09 |
ENSDART00000146001
|
wasf1
|
WAS protein family, member 1 |
| chr18_-_3166726 | 0.09 |
ENSDART00000165002
|
aqp11
|
aquaporin 11 |
| chr6_+_37752781 | 0.09 |
ENSDART00000154364
|
herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr10_+_22782522 | 0.09 |
ENSDART00000079498
ENSDART00000145558 |
si:ch211-237l4.6
|
si:ch211-237l4.6 |
| chr18_+_16986903 | 0.09 |
ENSDART00000142088
|
si:ch211-218c6.8
|
si:ch211-218c6.8 |
| chr3_-_13147310 | 0.09 |
ENSDART00000160840
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr22_+_36875400 | 0.08 |
ENSDART00000158293
|
mccc1
|
methylcrotonoyl-CoA carboxylase 1 (alpha) |
| chr6_-_48094342 | 0.08 |
ENSDART00000137458
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr3_-_5964557 | 0.08 |
ENSDART00000184738
|
BX284638.1
|
|
| chr25_+_35051656 | 0.08 |
ENSDART00000133379
|
hist2h3c
|
histone cluster 2, H3c |
| chr9_+_27411502 | 0.08 |
ENSDART00000143994
|
si:dkey-193n17.9
|
si:dkey-193n17.9 |
| chr13_+_12671513 | 0.08 |
ENSDART00000010517
|
eif4eb
|
eukaryotic translation initiation factor 4eb |
| chr14_-_8903435 | 0.08 |
ENSDART00000160584
|
zgc:153681
|
zgc:153681 |
| chr16_-_12095144 | 0.08 |
ENSDART00000145106
|
pex5
|
peroxisomal biogenesis factor 5 |
| chr12_-_48671612 | 0.08 |
ENSDART00000007202
|
zgc:92749
|
zgc:92749 |
| chr10_+_44903676 | 0.08 |
ENSDART00000158553
|
zgc:114173
|
zgc:114173 |
| chr15_-_6615555 | 0.08 |
ENSDART00000152725
|
atm
|
ATM serine/threonine kinase |
| chr23_-_2037566 | 0.08 |
ENSDART00000191312
ENSDART00000127443 |
prdm5
|
PR domain containing 5 |
| chr1_-_18803919 | 0.08 |
ENSDART00000020970
|
pgm2
|
phosphoglucomutase 2 |
| chr6_+_102506 | 0.07 |
ENSDART00000172678
|
ldlrb
|
low density lipoprotein receptor b |
| chr3_-_3327573 | 0.07 |
ENSDART00000136319
ENSDART00000166667 |
tmem184bb
|
transmembrane protein 184bb |
| chr5_+_24086227 | 0.07 |
ENSDART00000051549
ENSDART00000177458 ENSDART00000135934 |
tp53
|
tumor protein p53 |
| chr15_-_19250543 | 0.07 |
ENSDART00000092705
ENSDART00000138895 |
igsf9ba
|
immunoglobulin superfamily, member 9Ba |
| chr25_-_19433244 | 0.07 |
ENSDART00000154778
|
map1ab
|
microtubule-associated protein 1Ab |
| chr22_-_12693833 | 0.07 |
ENSDART00000129768
ENSDART00000044574 |
adarb1a
|
adenosine deaminase, RNA-specific, B1a |
| chr18_-_18942098 | 0.07 |
ENSDART00000100458
|
si:dkey-73n10.1
|
si:dkey-73n10.1 |
| chr13_+_4440869 | 0.07 |
ENSDART00000156323
|
tbxtb
|
T-box transcription factor Tb |
| chr11_+_45436703 | 0.07 |
ENSDART00000168295
ENSDART00000173293 |
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr18_-_2549198 | 0.07 |
ENSDART00000186516
|
CABZ01070631.1
|
|
| chr6_-_50704689 | 0.07 |
ENSDART00000074100
|
osgn1
|
oxidative stress induced growth inhibitor 1 |
| chr4_+_308697 | 0.06 |
ENSDART00000174021
ENSDART00000131419 ENSDART00000067489 |
si:ch211-51e12.7
|
si:ch211-51e12.7 |
| chr8_+_8927870 | 0.06 |
ENSDART00000081985
|
pim2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr8_-_44015210 | 0.06 |
ENSDART00000186879
ENSDART00000188965 ENSDART00000001313 ENSDART00000188902 ENSDART00000185935 ENSDART00000147869 |
rimbp2
rimbp2
|
RIMS binding protein 2 RIMS binding protein 2 |
| chr25_-_6389713 | 0.06 |
ENSDART00000083539
|
sin3aa
|
SIN3 transcription regulator family member Aa |
| chr7_-_18617187 | 0.06 |
ENSDART00000172419
|
si:ch211-119e14.1
|
si:ch211-119e14.1 |
| chr18_+_660578 | 0.06 |
ENSDART00000161203
|
CELF6
|
si:dkey-205h23.2 |
| chr12_+_25432627 | 0.06 |
ENSDART00000011662
|
ppm1bb
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Bb |
| chr19_-_10881141 | 0.06 |
ENSDART00000162793
|
psmd4a
|
proteasome 26S subunit, non-ATPase 4a |
| chr2_+_58877162 | 0.06 |
ENSDART00000122174
|
CABZ01085658.1
|
|
| chr3_+_17878466 | 0.05 |
ENSDART00000180218
|
dnajc7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr23_-_41651759 | 0.05 |
ENSDART00000146808
|
si:ch73-184c24.1
|
si:ch73-184c24.1 |
| chr16_+_6750289 | 0.05 |
ENSDART00000167736
|
znf236
|
zinc finger protein 236 |
| chr3_+_1492174 | 0.05 |
ENSDART00000112979
|
sox10
|
SRY (sex determining region Y)-box 10 |
| chr14_-_970853 | 0.05 |
ENSDART00000130801
|
acsl1b
|
acyl-CoA synthetase long chain family member 1b |
| chr7_-_69636502 | 0.05 |
ENSDART00000126739
|
tspan5a
|
tetraspanin 5a |
| chr6_-_218882 | 0.05 |
ENSDART00000008398
|
c1qtnf6b
|
C1q and TNF related 6b |
| chr3_-_62403550 | 0.05 |
ENSDART00000055055
|
sox8b
|
SRY (sex determining region Y)-box 8b |
| chr3_+_17878124 | 0.05 |
ENSDART00000166430
ENSDART00000163421 ENSDART00000121473 |
dnajc7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr16_-_44349845 | 0.05 |
ENSDART00000170932
|
rims2a
|
regulating synaptic membrane exocytosis 2a |
| chr25_+_35553542 | 0.05 |
ENSDART00000113723
|
spi1a
|
Spi-1 proto-oncogene a |
| chr23_+_2778813 | 0.05 |
ENSDART00000142621
|
top1
|
DNA topoisomerase I |
| chr25_-_36361697 | 0.05 |
ENSDART00000152388
|
si:ch211-113a14.22
|
si:ch211-113a14.22 |
| chr10_-_15644904 | 0.05 |
ENSDART00000138389
ENSDART00000101191 ENSDART00000186559 ENSDART00000122170 |
smc5
|
structural maintenance of chromosomes 5 |
| chr3_-_21062706 | 0.05 |
ENSDART00000155605
ENSDART00000153686 ENSDART00000157168 ENSDART00000156614 ENSDART00000155743 ENSDART00000156275 |
fam57ba
|
family with sequence similarity 57, member Ba |
| chr5_-_14500622 | 0.05 |
ENSDART00000099566
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr2_-_42091926 | 0.04 |
ENSDART00000142663
|
si:dkey-97a13.12
|
si:dkey-97a13.12 |
| chr18_-_49116382 | 0.04 |
ENSDART00000174157
|
BX663503.3
|
|
| chr24_-_39186185 | 0.04 |
ENSDART00000123019
ENSDART00000191114 |
nubp2
|
nucleotide binding protein 2 (MinD homolog, E. coli) |
| chr11_-_40519886 | 0.04 |
ENSDART00000172819
|
miip
|
migration and invasion inhibitory protein |
| chr7_-_6466119 | 0.04 |
ENSDART00000173138
|
zgc:112234
|
zgc:112234 |
| chr11_+_18612166 | 0.04 |
ENSDART00000162694
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr7_+_57108823 | 0.04 |
ENSDART00000184943
ENSDART00000055956 |
enosf1
|
enolase superfamily member 1 |
| chr4_+_29967255 | 0.04 |
ENSDART00000150383
|
znf1128
|
zinc finger protein 1128 |
| chr10_-_35236949 | 0.04 |
ENSDART00000145804
|
ypel2a
|
yippee-like 2a |
| chr16_+_45930962 | 0.04 |
ENSDART00000124689
ENSDART00000041811 |
otud7b
|
OTU deubiquitinase 7B |
| chr6_+_56147812 | 0.04 |
ENSDART00000150219
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr1_+_54069450 | 0.04 |
ENSDART00000108601
ENSDART00000187878 |
dcaf15
|
DDB1 and CUL4 associated factor 15 |
| chr7_-_31794476 | 0.04 |
ENSDART00000142385
|
nap1l4b
|
nucleosome assembly protein 1-like 4b |
| chr5_+_42092227 | 0.04 |
ENSDART00000097583
ENSDART00000171678 |
ubb
|
ubiquitin B |
| chr21_-_12119711 | 0.04 |
ENSDART00000131538
|
celf4
|
CUGBP, Elav-like family member 4 |
| chr13_-_30645965 | 0.04 |
ENSDART00000109307
|
zcchc24
|
zinc finger, CCHC domain containing 24 |
| chr13_+_42602406 | 0.04 |
ENSDART00000133388
ENSDART00000147996 |
mlh1
|
mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli) |
| chr11_+_77526 | 0.04 |
ENSDART00000193521
|
CABZ01072242.1
|
|
| chr1_-_20271138 | 0.04 |
ENSDART00000185931
|
ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr11_-_42554290 | 0.04 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr13_-_856525 | 0.03 |
ENSDART00000143356
|
TMEM14A
|
zgc:163080 |
| chr16_-_52646789 | 0.03 |
ENSDART00000035761
|
ubr5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr6_+_3710865 | 0.03 |
ENSDART00000170781
|
phospho2
|
phosphatase, orphan 2 |
| chr11_+_440305 | 0.03 |
ENSDART00000082517
|
rab43
|
RAB43, member RAS oncogene family |
| chr4_-_240737 | 0.03 |
ENSDART00000166186
|
RERG
|
si:cabz01085950.1 |
| chr15_+_26941063 | 0.03 |
ENSDART00000149957
|
bcas3
|
breast carcinoma amplified sequence 3 |
| chr23_+_37185247 | 0.03 |
ENSDART00000146269
|
vwa5b1
|
von Willebrand factor A domain containing 5B1 |
| chr19_-_10881486 | 0.03 |
ENSDART00000168852
ENSDART00000160438 |
PSMD4 (1 of many)
psmd4a
|
proteasome 26S subunit, non-ATPase 4 proteasome 26S subunit, non-ATPase 4a |
| chr1_-_9644630 | 0.03 |
ENSDART00000123725
ENSDART00000161164 |
ugt5b3
|
UDP glucuronosyltransferase 5 family, polypeptide B3 |
| chr22_-_4439311 | 0.03 |
ENSDART00000169317
|
uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr17_-_31221655 | 0.03 |
ENSDART00000145010
ENSDART00000137834 ENSDART00000132858 |
pcmtl
|
l-isoaspartyl protein carboxyl methyltransferase, like |
| chr21_+_5888641 | 0.03 |
ENSDART00000091331
|
prodha
|
proline dehydrogenase (oxidase) 1a |
| chr15_-_31177324 | 0.03 |
ENSDART00000008854
|
wsb1
|
WD repeat and SOCS box containing 1 |
| chr3_-_1146497 | 0.03 |
ENSDART00000149061
|
si:ch73-211l13.2
|
si:ch73-211l13.2 |
| chr23_+_14590483 | 0.03 |
ENSDART00000088359
ENSDART00000184868 |
si:rp71-79p20.2
|
si:rp71-79p20.2 |
| chr6_+_8622228 | 0.03 |
ENSDART00000110470
|
tspeara
|
thrombospondin-type laminin G domain and EAR repeats a |
| chr13_-_856701 | 0.03 |
ENSDART00000140423
|
TMEM14A
|
zgc:163080 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ybx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0032877 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.1 | 0.4 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.1 | 0.3 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 0.2 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 0.2 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.1 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.0 | 0.2 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.2 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.0 | 0.2 | GO:0010896 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.2 | GO:2000051 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:0050428 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.3 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.0 | 0.1 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.0 | 0.1 | GO:0032197 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.0 | 0.2 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.4 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.2 | GO:0055129 | arginine catabolic process(GO:0006527) proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.3 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.2 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.5 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 0.2 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.0 | GO:0051026 | chiasma assembly(GO:0051026) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.1 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.5 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.2 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.1 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.2 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.8 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.3 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |