Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
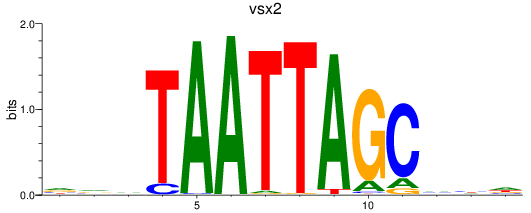
Results for vsx2
Z-value: 0.37
Transcription factors associated with vsx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
vsx2
|
ENSDARG00000005574 | visual system homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| vsx2 | dr11_v1_chr17_-_31659670_31659670 | -0.80 | 6.2e-05 | Click! |
Activity profile of vsx2 motif
Sorted Z-values of vsx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_35633827 | 1.39 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr6_+_28208973 | 1.21 |
ENSDART00000171216
ENSDART00000171377 ENSDART00000167389 ENSDART00000166988 |
LSM2 (1 of many)
|
si:ch73-14h10.2 |
| chr2_-_15324837 | 0.93 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr20_+_29209767 | 0.81 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr20_+_29209926 | 0.75 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr7_+_22313533 | 0.72 |
ENSDART00000123457
|
TMEM102
|
si:dkey-11f12.2 |
| chr20_+_29209615 | 0.69 |
ENSDART00000062350
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr13_-_24717618 | 0.69 |
ENSDART00000172156
|
erlin1
|
ER lipid raft associated 1 |
| chr16_+_39159752 | 0.68 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr22_-_21897203 | 0.64 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr12_-_35386910 | 0.62 |
ENSDART00000153453
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr13_-_24717365 | 0.60 |
ENSDART00000137934
ENSDART00000003922 |
erlin1
|
ER lipid raft associated 1 |
| chr21_-_32781612 | 0.58 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr6_-_40713183 | 0.57 |
ENSDART00000157113
ENSDART00000154810 ENSDART00000153702 |
si:ch211-157b11.12
|
si:ch211-157b11.12 |
| chr19_+_15441022 | 0.47 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr13_+_38817871 | 0.47 |
ENSDART00000187708
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr11_+_18873619 | 0.47 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr24_+_19415124 | 0.44 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr19_+_15440841 | 0.44 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr14_+_21820034 | 0.43 |
ENSDART00000122739
|
ctbp1
|
C-terminal binding protein 1 |
| chr23_-_33709964 | 0.42 |
ENSDART00000143333
ENSDART00000130338 |
pou6f1
|
POU class 6 homeobox 1 |
| chr2_-_26596794 | 0.41 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr8_-_15129573 | 0.40 |
ENSDART00000142358
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr20_-_20270191 | 0.39 |
ENSDART00000009356
|
ppp2r5ea
|
protein phosphatase 2, regulatory subunit B', epsilon isoform a |
| chr24_-_25004553 | 0.37 |
ENSDART00000080997
ENSDART00000136860 |
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr1_+_9290103 | 0.37 |
ENSDART00000055009
|
uncx4.1
|
Unc4.1 homeobox (C. elegans) |
| chr24_+_39518774 | 0.31 |
ENSDART00000132939
|
dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr8_-_23776399 | 0.30 |
ENSDART00000114800
|
INAVA
|
si:ch211-163l21.4 |
| chr7_+_36898850 | 0.30 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr13_-_36798204 | 0.29 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr21_-_13662237 | 0.29 |
ENSDART00000091647
ENSDART00000151547 |
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr15_-_9272328 | 0.28 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr3_-_43356082 | 0.26 |
ENSDART00000171213
|
uncx
|
UNC homeobox |
| chr20_-_37813863 | 0.24 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr9_-_50001606 | 0.22 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr9_-_3934963 | 0.22 |
ENSDART00000062336
|
ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr5_-_25733745 | 0.21 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr6_-_39198912 | 0.18 |
ENSDART00000077938
|
c1galt1b
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1b |
| chr6_-_39199070 | 0.17 |
ENSDART00000131793
|
c1galt1b
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1b |
| chr5_+_5398966 | 0.16 |
ENSDART00000139553
|
mapkap1
|
mitogen-activated protein kinase associated protein 1 |
| chr4_+_3980247 | 0.15 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr12_+_30367371 | 0.13 |
ENSDART00000153364
|
ccdc186
|
si:ch211-225b10.4 |
| chr5_-_15851953 | 0.12 |
ENSDART00000173101
|
si:dkey-1k23.3
|
si:dkey-1k23.3 |
| chr14_+_23874062 | 0.12 |
ENSDART00000172149
|
sh3rf2
|
SH3 domain containing ring finger 2 |
| chr2_-_35566938 | 0.11 |
ENSDART00000029006
ENSDART00000077178 ENSDART00000125298 |
tnn
|
tenascin N |
| chr23_-_24483311 | 0.11 |
ENSDART00000185793
ENSDART00000109248 |
spen
|
spen family transcriptional repressor |
| chr14_-_7207961 | 0.11 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr4_+_21129752 | 0.10 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr12_+_30367079 | 0.10 |
ENSDART00000190112
|
ccdc186
|
si:ch211-225b10.4 |
| chr16_-_22930925 | 0.10 |
ENSDART00000133819
|
si:dkey-246i14.3
|
si:dkey-246i14.3 |
| chr20_-_9436521 | 0.10 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr24_+_1023839 | 0.09 |
ENSDART00000082526
|
zgc:111976
|
zgc:111976 |
| chr14_+_23717165 | 0.07 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr19_+_2631565 | 0.07 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr25_-_3836597 | 0.07 |
ENSDART00000115154
|
si:ch211-247i17.1
|
si:ch211-247i17.1 |
| chr5_+_38900249 | 0.07 |
ENSDART00000097856
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr5_-_16996482 | 0.06 |
ENSDART00000144501
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr21_+_28958471 | 0.06 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr21_-_13661631 | 0.06 |
ENSDART00000184408
|
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr12_-_19862912 | 0.05 |
ENSDART00000145788
|
shisa9a
|
shisa family member 9a |
| chr21_+_25231160 | 0.05 |
ENSDART00000063089
ENSDART00000139127 |
gng8
|
guanine nucleotide binding protein (G protein), gamma 8 |
| chr19_-_19379084 | 0.05 |
ENSDART00000165206
|
smarcc1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
| chr25_+_6471401 | 0.04 |
ENSDART00000132927
|
snx33
|
sorting nexin 33 |
| chr23_-_24195519 | 0.04 |
ENSDART00000112370
ENSDART00000180377 |
ano11
|
anoctamin 11 |
| chr23_-_24195723 | 0.04 |
ENSDART00000145489
|
ano11
|
anoctamin 11 |
| chr22_+_9003090 | 0.04 |
ENSDART00000106414
|
rnh1
|
ribonuclease/angiogenin inhibitor 1 |
| chr12_-_35095414 | 0.03 |
ENSDART00000153229
|
si:dkey-21e13.3
|
si:dkey-21e13.3 |
| chr11_+_18873113 | 0.02 |
ENSDART00000103969
ENSDART00000103968 |
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr9_+_50001746 | 0.02 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr18_+_28106139 | 0.02 |
ENSDART00000089615
|
kiaa1549lb
|
KIAA1549-like b |
| chr19_-_5265155 | 0.01 |
ENSDART00000145003
|
prf1.3
|
perforin 1.3 |
| chr1_+_55755304 | 0.00 |
ENSDART00000144983
|
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr1_-_11372456 | 0.00 |
ENSDART00000144164
ENSDART00000141238 |
sdk1b
|
sidekick cell adhesion molecule 1b |
| chr9_-_23152092 | 0.00 |
ENSDART00000180155
ENSDART00000186935 |
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr3_-_21397304 | 0.00 |
ENSDART00000189794
|
CT573446.1
|
|
| chr6_-_12275836 | 0.00 |
ENSDART00000189980
|
pkp4
|
plakophilin 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of vsx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 1.3 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.4 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.9 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.2 | GO:0090134 | establishment or maintenance of cytoskeleton polarity(GO:0030952) mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.4 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.4 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.4 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.6 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.6 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.4 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 1.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.4 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.9 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |