Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
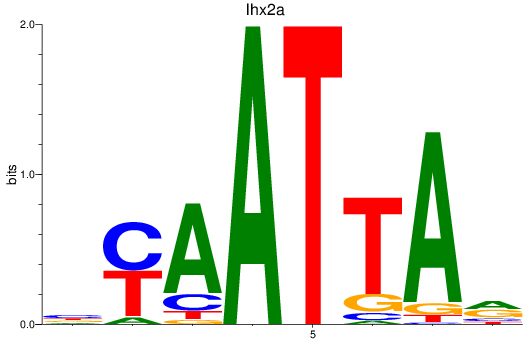
Results for vsx1_shox_shox2_uncx4.1_lhx2a
Z-value: 1.15
Transcription factors associated with vsx1_shox_shox2_uncx4.1_lhx2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
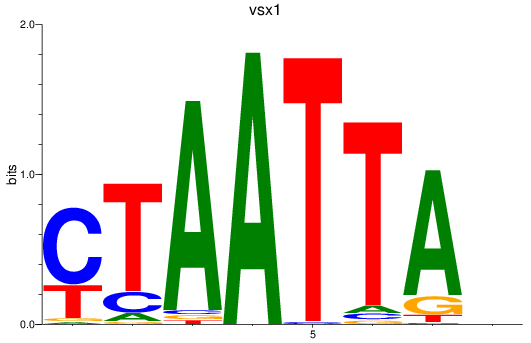
vsx1
|
ENSDARG00000056292 | visual system homeobox 1 homolog, chx10-like |
|
vsx1
|
ENSDARG00000109766 | visual system homeobox 1 homolog, chx10-like |
|
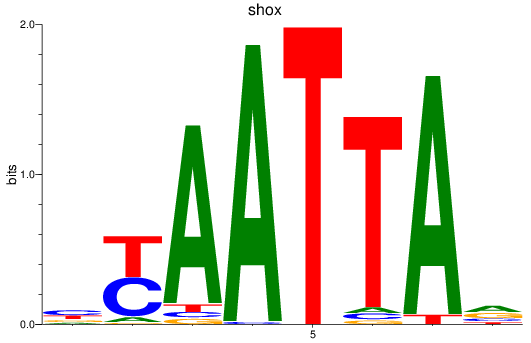
shox
|
ENSDARG00000025891 | short stature homeobox |
|
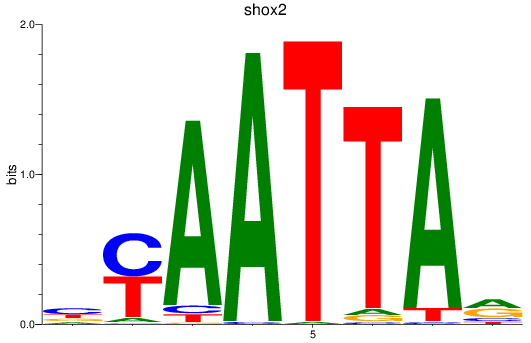
shox2
|
ENSDARG00000075713 | short stature homeobox 2 |
|
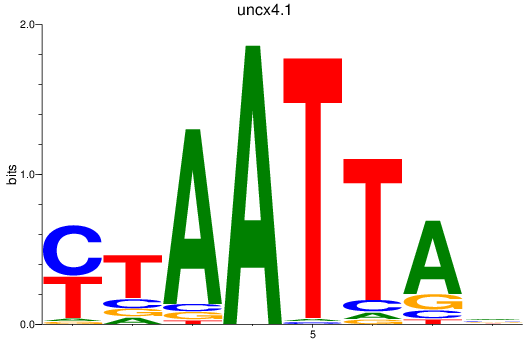
uncx4.1
|
ENSDARG00000037760 | Unc4.1 homeobox (C. elegans) |
|
lhx2a
|
ENSDARG00000037964 | LIM homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lhx2a | dr11_v1_chr21_-_8422351_8422363 | -0.98 | 1.7e-12 | Click! |
| uncx4.1 | dr11_v1_chr1_+_9290103_9290103 | 0.87 | 2.3e-06 | Click! |
| shox | dr11_v1_chr9_+_34641237_34641237 | -0.86 | 5.6e-06 | Click! |
| vsx1 | dr11_v1_chr17_-_21066075_21066105 | -0.61 | 7.7e-03 | Click! |
| shox2 | dr11_v1_chr15_-_2188332_2188422 | -0.19 | 4.4e-01 | Click! |
Activity profile of vsx1_shox_shox2_uncx4.1_lhx2a motif
Sorted Z-values of vsx1_shox_shox2_uncx4.1_lhx2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_21362071 | 10.19 |
ENSDART00000125167
|
avd
|
avidin |
| chr10_-_21362320 | 9.54 |
ENSDART00000189789
|
avd
|
avidin |
| chr18_-_40708537 | 9.27 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr21_+_25777425 | 9.20 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr11_-_6452444 | 8.76 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr8_+_45334255 | 7.14 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr16_+_29509133 | 7.11 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr20_-_23426339 | 6.83 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr9_-_35633827 | 6.75 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr11_-_44801968 | 6.67 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr5_+_37903790 | 6.37 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr10_-_25217347 | 5.74 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr10_-_34915886 | 5.10 |
ENSDART00000141201
ENSDART00000002166 |
ccna1
|
cyclin A1 |
| chr10_-_34916208 | 4.87 |
ENSDART00000187371
|
ccna1
|
cyclin A1 |
| chr10_-_34002185 | 4.47 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr2_+_41526904 | 4.30 |
ENSDART00000127520
|
acvr1l
|
activin A receptor, type 1 like |
| chr2_+_6253246 | 4.29 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr1_-_18811517 | 4.28 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr11_-_1550709 | 4.27 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr1_-_55248496 | 4.17 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr24_+_12835935 | 4.03 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr2_-_38284648 | 3.99 |
ENSDART00000148281
ENSDART00000132621 |
si:ch211-14a17.7
|
si:ch211-14a17.7 |
| chr8_-_20230559 | 3.88 |
ENSDART00000193677
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr14_+_34490445 | 3.86 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr17_+_16046132 | 3.82 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr2_-_38287987 | 3.78 |
ENSDART00000185329
ENSDART00000061677 |
si:ch211-14a17.6
|
si:ch211-14a17.6 |
| chr17_+_16046314 | 3.76 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr20_-_6532462 | 3.66 |
ENSDART00000054653
|
mcm3l
|
MCM3 minichromosome maintenance deficient 3 (S. cerevisiae), like |
| chr11_+_18183220 | 3.65 |
ENSDART00000113468
|
LO018315.10
|
|
| chr8_+_11425048 | 3.38 |
ENSDART00000018739
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr8_-_20230802 | 3.25 |
ENSDART00000063400
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr7_-_48263516 | 3.18 |
ENSDART00000006619
ENSDART00000142370 ENSDART00000148273 ENSDART00000147968 |
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr16_+_39159752 | 3.10 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr2_-_15324837 | 3.02 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr16_-_17197546 | 2.85 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr2_-_26596794 | 2.79 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr4_+_9467049 | 2.77 |
ENSDART00000012659
|
zgc:55888
|
zgc:55888 |
| chr6_+_21001264 | 2.66 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr5_-_68333081 | 2.63 |
ENSDART00000168786
|
h1m
|
linker histone H1M |
| chr3_+_28860283 | 2.57 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr8_-_25034411 | 2.48 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr13_+_38817871 | 2.44 |
ENSDART00000187708
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr5_+_60590796 | 2.43 |
ENSDART00000159859
|
tmem132e
|
transmembrane protein 132E |
| chr23_-_33709964 | 2.31 |
ENSDART00000143333
ENSDART00000130338 |
pou6f1
|
POU class 6 homeobox 1 |
| chr21_+_34088110 | 2.22 |
ENSDART00000145123
ENSDART00000029599 ENSDART00000147519 |
mtmr1b
|
myotubularin related protein 1b |
| chr1_+_35985813 | 2.18 |
ENSDART00000179634
ENSDART00000139636 ENSDART00000175902 |
zgc:152968
|
zgc:152968 |
| chr4_+_13586689 | 2.17 |
ENSDART00000067161
ENSDART00000138201 |
tnpo3
|
transportin 3 |
| chr11_+_18873619 | 2.13 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr16_-_29387215 | 2.12 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr14_+_35428152 | 2.01 |
ENSDART00000172597
|
sytl4
|
synaptotagmin-like 4 |
| chr6_+_40922572 | 1.97 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr6_-_12172424 | 1.92 |
ENSDART00000109344
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr20_-_45060241 | 1.92 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr23_+_2728095 | 1.89 |
ENSDART00000066086
|
zgc:114123
|
zgc:114123 |
| chr3_-_32337653 | 1.89 |
ENSDART00000156918
ENSDART00000156551 |
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr21_-_32060993 | 1.75 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr15_-_25099679 | 1.73 |
ENSDART00000154628
|
rflnb
|
refilin B |
| chr12_+_22580579 | 1.72 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr24_+_19415124 | 1.72 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr7_+_24023653 | 1.70 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr14_+_8940326 | 1.70 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr24_-_25144441 | 1.70 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr10_+_43039947 | 1.67 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr24_-_2450597 | 1.67 |
ENSDART00000188080
ENSDART00000093331 |
rreb1a
|
ras responsive element binding protein 1a |
| chr20_-_40758410 | 1.66 |
ENSDART00000183031
|
cx34.5
|
connexin 34.5 |
| chr15_-_2519640 | 1.66 |
ENSDART00000047013
|
srprb
|
signal recognition particle receptor, B subunit |
| chr6_-_40922971 | 1.61 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr13_-_25720876 | 1.59 |
ENSDART00000142404
ENSDART00000146487 |
si:dkey-192p21.6
|
si:dkey-192p21.6 |
| chr10_-_21545091 | 1.58 |
ENSDART00000029122
ENSDART00000132207 |
zgc:165539
|
zgc:165539 |
| chr10_+_11261576 | 1.58 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr8_+_11325310 | 1.57 |
ENSDART00000142577
|
fxn
|
frataxin |
| chr4_-_77125693 | 1.56 |
ENSDART00000174256
|
CU467646.3
|
|
| chr19_+_15441022 | 1.50 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr17_-_30635298 | 1.50 |
ENSDART00000155478
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr17_-_41798856 | 1.46 |
ENSDART00000156031
ENSDART00000192801 ENSDART00000180172 ENSDART00000084745 ENSDART00000175577 |
ralgapa2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr20_+_32523576 | 1.45 |
ENSDART00000147319
|
scml4
|
Scm polycomb group protein like 4 |
| chr20_+_29209926 | 1.45 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr5_+_57924611 | 1.42 |
ENSDART00000050949
|
btg4
|
B-cell translocation gene 4 |
| chr13_+_38814521 | 1.39 |
ENSDART00000110976
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr3_-_26183699 | 1.39 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr20_+_29209767 | 1.38 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr17_-_24575893 | 1.37 |
ENSDART00000141914
|
aftphb
|
aftiphilin b |
| chr15_-_30857350 | 1.36 |
ENSDART00000138988
|
akap1b
|
A kinase (PRKA) anchor protein 1b |
| chr20_-_37813863 | 1.35 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr11_+_18130300 | 1.33 |
ENSDART00000169146
|
zgc:175135
|
zgc:175135 |
| chr3_-_30488063 | 1.32 |
ENSDART00000055393
ENSDART00000151367 |
med25
|
mediator complex subunit 25 |
| chr8_-_23780334 | 1.30 |
ENSDART00000145179
ENSDART00000145894 |
zgc:195245
|
zgc:195245 |
| chr22_-_20924564 | 1.28 |
ENSDART00000100642
ENSDART00000032770 |
ell
|
elongation factor RNA polymerase II |
| chr22_-_17653143 | 1.27 |
ENSDART00000089171
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr19_+_15440841 | 1.26 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr10_-_32494499 | 1.24 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr21_+_34088377 | 1.24 |
ENSDART00000170070
|
mtmr1b
|
myotubularin related protein 1b |
| chr11_+_18157260 | 1.22 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr21_+_15592426 | 1.22 |
ENSDART00000138207
|
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr13_-_31017960 | 1.21 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr17_+_37227936 | 1.21 |
ENSDART00000076009
|
hadhab
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha b |
| chr5_-_25733745 | 1.21 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr23_-_17003533 | 1.20 |
ENSDART00000080545
|
dnmt3bb.2
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.2 |
| chr4_-_77116266 | 1.19 |
ENSDART00000174249
|
CU467646.2
|
|
| chr14_-_33945692 | 1.18 |
ENSDART00000168546
ENSDART00000189778 |
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr11_-_35171162 | 1.16 |
ENSDART00000017393
|
traip
|
TRAF-interacting protein |
| chr4_-_77130289 | 1.14 |
ENSDART00000174380
|
CU467646.7
|
|
| chr1_-_40102836 | 1.12 |
ENSDART00000147317
|
cntf
|
ciliary neurotrophic factor |
| chr25_+_18475032 | 1.12 |
ENSDART00000073564
|
tes
|
testis derived transcript (3 LIM domains) |
| chr25_-_3867990 | 1.10 |
ENSDART00000075663
|
cracr2b
|
calcium release activated channel regulator 2B |
| chr20_+_29209615 | 1.10 |
ENSDART00000062350
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr5_+_25733774 | 1.08 |
ENSDART00000137088
ENSDART00000098467 |
abhd17b
|
abhydrolase domain containing 17B |
| chr20_-_28800999 | 1.06 |
ENSDART00000049462
|
rab15
|
RAB15, member RAS oncogene family |
| chr14_-_33481428 | 1.06 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr2_-_7246848 | 1.04 |
ENSDART00000146434
|
zgc:153115
|
zgc:153115 |
| chr8_-_15129573 | 1.04 |
ENSDART00000142358
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr3_+_56366395 | 1.03 |
ENSDART00000154367
|
cacng5b
|
calcium channel, voltage-dependent, gamma subunit 5b |
| chr18_+_15644559 | 1.03 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr19_+_21362553 | 1.03 |
ENSDART00000122002
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr3_-_26244256 | 1.01 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr1_-_6028876 | 1.01 |
ENSDART00000168117
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr8_+_18010568 | 1.01 |
ENSDART00000121984
|
ssbp3b
|
single stranded DNA binding protein 3b |
| chr13_-_31008275 | 1.00 |
ENSDART00000139394
|
wdfy4
|
WDFY family member 4 |
| chr2_-_38363017 | 0.99 |
ENSDART00000088026
|
prmt5
|
protein arginine methyltransferase 5 |
| chr10_-_32494304 | 0.99 |
ENSDART00000028161
|
uvrag
|
UV radiation resistance associated gene |
| chr21_-_39177564 | 0.99 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr15_+_17258246 | 0.96 |
ENSDART00000101707
|
dhx40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr8_-_32385989 | 0.95 |
ENSDART00000143716
ENSDART00000098850 |
lipg
|
lipase, endothelial |
| chr24_-_34680956 | 0.94 |
ENSDART00000171009
|
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr8_-_19467011 | 0.92 |
ENSDART00000162010
|
zgc:92140
|
zgc:92140 |
| chr24_-_30862168 | 0.92 |
ENSDART00000168540
|
ptbp2a
|
polypyrimidine tract binding protein 2a |
| chr25_-_27621268 | 0.92 |
ENSDART00000146205
ENSDART00000073511 |
hyal6
|
hyaluronoglucosaminidase 6 |
| chr21_-_28640316 | 0.92 |
ENSDART00000128237
|
nrg2a
|
neuregulin 2a |
| chr10_-_8053753 | 0.91 |
ENSDART00000162289
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr13_+_2625150 | 0.91 |
ENSDART00000164177
|
plpp4
|
phospholipid phosphatase 4 |
| chr14_+_22113331 | 0.91 |
ENSDART00000109759
|
tmx2a
|
thioredoxin-related transmembrane protein 2a |
| chr9_-_50001606 | 0.91 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr17_-_11439815 | 0.89 |
ENSDART00000130105
|
psma3
|
proteasome subunit alpha 3 |
| chr2_-_15318786 | 0.89 |
ENSDART00000135851
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr7_-_51773166 | 0.89 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr2_-_49031303 | 0.89 |
ENSDART00000143471
|
cdc34b
|
cell division cycle 34 homolog (S. cerevisiae) b |
| chr3_-_26787430 | 0.88 |
ENSDART00000087047
|
rab40c
|
RAB40c, member RAS oncogene family |
| chr17_-_49412313 | 0.88 |
ENSDART00000152100
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr12_-_26153101 | 0.87 |
ENSDART00000076051
|
opn4b
|
opsin 4b |
| chr1_+_513986 | 0.86 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr20_-_52902693 | 0.86 |
ENSDART00000166115
ENSDART00000161050 |
ctsbb
|
cathepsin Bb |
| chr8_-_7232413 | 0.85 |
ENSDART00000092426
|
grip2a
|
glutamate receptor interacting protein 2a |
| chr25_+_31405266 | 0.85 |
ENSDART00000103395
|
tnnt3a
|
troponin T type 3a (skeletal, fast) |
| chr1_+_24387659 | 0.85 |
ENSDART00000130356
|
qdprb2
|
quinoid dihydropteridine reductase b2 |
| chr15_-_26931541 | 0.85 |
ENSDART00000027563
|
ccdc9
|
coiled-coil domain containing 9 |
| chr17_-_6613458 | 0.83 |
ENSDART00000175024
|
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr15_-_9272328 | 0.83 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr8_-_25033681 | 0.83 |
ENSDART00000003493
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr24_+_26134209 | 0.82 |
ENSDART00000038824
|
tmtopsb
|
teleost multiple tissue opsin b |
| chr20_-_9436521 | 0.79 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr17_+_19630272 | 0.78 |
ENSDART00000104895
|
rgs7a
|
regulator of G protein signaling 7a |
| chr3_-_43356082 | 0.78 |
ENSDART00000171213
|
uncx
|
UNC homeobox |
| chr8_+_50953776 | 0.78 |
ENSDART00000013870
|
zgc:56596
|
zgc:56596 |
| chr15_-_18115540 | 0.76 |
ENSDART00000131639
ENSDART00000047902 |
arcn1b
|
archain 1b |
| chr11_+_31864921 | 0.76 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr13_-_36911118 | 0.76 |
ENSDART00000048739
|
trim9
|
tripartite motif containing 9 |
| chr19_-_19871211 | 0.76 |
ENSDART00000170980
|
evx1
|
even-skipped homeobox 1 |
| chr12_-_18577983 | 0.75 |
ENSDART00000193262
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr15_+_12429206 | 0.75 |
ENSDART00000168997
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr7_+_24645186 | 0.75 |
ENSDART00000150118
ENSDART00000150233 ENSDART00000087691 |
gba2
|
glucosidase, beta (bile acid) 2 |
| chr19_+_7549854 | 0.75 |
ENSDART00000138866
ENSDART00000151758 |
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr8_+_25034544 | 0.74 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr16_+_20161805 | 0.74 |
ENSDART00000192146
|
c16h2orf66
|
chromosome 16 C2orf66 homolog |
| chr2_+_37227011 | 0.74 |
ENSDART00000126587
ENSDART00000084958 |
samd7
|
sterile alpha motif domain containing 7 |
| chr19_-_25119443 | 0.74 |
ENSDART00000148953
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr18_+_50880096 | 0.73 |
ENSDART00000169782
|
si:ch1073-450f2.1
|
si:ch1073-450f2.1 |
| chr9_-_746317 | 0.72 |
ENSDART00000129632
ENSDART00000130720 |
usp37
|
ubiquitin specific peptidase 37 |
| chr19_-_43750389 | 0.72 |
ENSDART00000147328
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr5_-_30074332 | 0.72 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr10_-_13343831 | 0.72 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr3_+_18807006 | 0.71 |
ENSDART00000180091
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr9_-_51436377 | 0.71 |
ENSDART00000006612
|
tbr1b
|
T-box, brain, 1b |
| chr22_+_17828267 | 0.71 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr12_+_38563373 | 0.70 |
ENSDART00000134670
ENSDART00000193668 |
ttyh2
|
tweety family member 2 |
| chr7_+_29167744 | 0.70 |
ENSDART00000076345
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr18_-_15551360 | 0.70 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr2_+_50608099 | 0.69 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr4_+_3980247 | 0.69 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr2_+_51783120 | 0.69 |
ENSDART00000177559
|
crygn1
|
crystallin, gamma N1 |
| chr13_-_8692860 | 0.68 |
ENSDART00000058107
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr12_+_48803098 | 0.67 |
ENSDART00000074768
|
ppifb
|
peptidylprolyl isomerase Fb |
| chr20_+_28803977 | 0.67 |
ENSDART00000153351
ENSDART00000038149 |
fntb
|
farnesyltransferase, CAAX box, beta |
| chr7_+_28612671 | 0.66 |
ENSDART00000019991
|
slc7a6os
|
solute carrier family 7, member 6 opposite strand |
| chr25_+_35891342 | 0.65 |
ENSDART00000147093
|
lsm14aa
|
LSM14A mRNA processing body assembly factor a |
| chr9_+_50001746 | 0.64 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr8_-_50888806 | 0.64 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr24_-_38110779 | 0.64 |
ENSDART00000147783
|
crp
|
c-reactive protein, pentraxin-related |
| chr19_+_7152966 | 0.63 |
ENSDART00000080348
|
brd2a
|
bromodomain containing 2a |
| chr13_+_38430466 | 0.63 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr14_+_34492288 | 0.62 |
ENSDART00000144301
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr25_-_2723682 | 0.62 |
ENSDART00000113382
|
adpgk
|
ADP-dependent glucokinase |
| chr10_-_33297864 | 0.62 |
ENSDART00000163360
|
PRDM15
|
PR/SET domain 15 |
| chr20_-_28642061 | 0.60 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr2_-_57076687 | 0.60 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr12_-_33817114 | 0.60 |
ENSDART00000161265
|
twnk
|
twinkle mtDNA helicase |
| chr16_+_25116827 | 0.59 |
ENSDART00000163244
|
si:ch211-261d7.6
|
si:ch211-261d7.6 |
| chr22_-_20166660 | 0.59 |
ENSDART00000085913
ENSDART00000188241 |
btbd2a
|
BTB (POZ) domain containing 2a |
| chr16_-_54919260 | 0.58 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr10_+_17371356 | 0.58 |
ENSDART00000122663
|
sppl3
|
signal peptide peptidase 3 |
| chr14_+_30795559 | 0.58 |
ENSDART00000006132
|
cfl1
|
cofilin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of vsx1_shox_shox2_uncx4.1_lhx2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 9.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 1.5 | 4.5 | GO:0072111 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 1.1 | 4.3 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 1.0 | 4.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.8 | 3.1 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.7 | 12.4 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.6 | 3.2 | GO:0051148 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.6 | 1.7 | GO:1900157 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.5 | 3.7 | GO:1902292 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.5 | 5.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.3 | 6.6 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.3 | 1.7 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.2 | 0.7 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.2 | 1.0 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.2 | 1.2 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.2 | 1.9 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.2 | 0.9 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.2 | 1.7 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.2 | 1.3 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.2 | 0.6 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 0.8 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.2 | 1.1 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.2 | 9.0 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.2 | 1.9 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.2 | 0.7 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.2 | 2.0 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 3.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 2.6 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 4.5 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 0.9 | GO:0003222 | ventricular trabecula myocardium morphogenesis(GO:0003222) trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) skin epidermis development(GO:0098773) |
| 0.1 | 1.3 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.6 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 0.5 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.1 | 1.0 | GO:2001287 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 1.7 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.1 | 0.6 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.1 | 1.0 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 1.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.3 | GO:0009085 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.1 | 0.7 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.5 | GO:0006660 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 1.6 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.4 | GO:0042182 | ketone catabolic process(GO:0042182) |
| 0.1 | 0.5 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 4.9 | GO:0018198 | peptidyl-cysteine modification(GO:0018198) |
| 0.1 | 0.5 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.4 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 8.9 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.1 | 1.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.4 | GO:0071422 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.1 | 0.8 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 0.8 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.1 | 1.7 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.7 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.7 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 0.5 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 2.7 | GO:0060968 | regulation of gene silencing(GO:0060968) |
| 0.1 | 0.3 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.1 | 1.7 | GO:0072599 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.1 | 0.7 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.1 | 0.6 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 1.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.5 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.1 | 0.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 1.0 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.1 | 0.3 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.3 | GO:0016246 | RNA interference(GO:0016246) |
| 0.1 | 1.7 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.7 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.1 | 1.0 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.1 | 0.9 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.3 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.1 | 1.0 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.4 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.1 | 0.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.2 | GO:1902260 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 0.4 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 4.3 | GO:0048846 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.1 | 0.9 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.5 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 2.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.4 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 1.0 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.6 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 2.7 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 1.7 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.9 | GO:0033138 | regulation of peptidyl-serine phosphorylation(GO:0033135) positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.2 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.0 | 2.4 | GO:0006497 | protein lipidation(GO:0006497) |
| 0.0 | 0.4 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 1.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.8 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.0 | 0.2 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.1 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.6 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.7 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.5 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.3 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.8 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.0 | GO:0043090 | amino acid import(GO:0043090) L-amino acid import(GO:0043092) |
| 0.0 | 0.1 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.0 | 1.2 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.5 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.3 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.0 | 1.2 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.5 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 | 0.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0043490 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:1900120 | microglial cell activation(GO:0001774) regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.0 | 0.2 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0070254 | mucus secretion(GO:0070254) regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 1.5 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 0.4 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 | 0.2 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.5 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.1 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.3 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.0 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 0.2 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.5 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.4 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 0.1 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.5 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.8 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 0.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 1.7 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 1.2 | GO:0048545 | response to steroid hormone(GO:0048545) |
| 0.0 | 0.0 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.2 | GO:0051492 | regulation of stress fiber assembly(GO:0051492) |
| 0.0 | 0.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) synaptic vesicle budding(GO:0070142) |
| 0.0 | 0.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 1.0 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.1 | GO:0031060 | regulation of histone methylation(GO:0031060) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 10.0 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 1.5 | 4.5 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.9 | 4.3 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.4 | 1.1 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.3 | 3.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.3 | 1.0 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.2 | 7.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 3.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 1.4 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.2 | 0.7 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.2 | 5.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 1.2 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 1.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.8 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 1.7 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 14.1 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 0.6 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.4 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 1.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.4 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.3 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 1.7 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 2.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.8 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.6 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.9 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.3 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.5 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 3.1 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.2 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.0 | 1.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.7 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 1.8 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 2.8 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 2.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 6.6 | GO:0000323 | lytic vacuole(GO:0000323) |
| 0.0 | 0.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 9.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.6 | GO:0000313 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 1.0 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 2.7 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 1.5 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 3.5 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.1 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 19.7 | GO:0009374 | biotin binding(GO:0009374) |
| 0.7 | 12.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.7 | 2.8 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.6 | 1.7 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.5 | 7.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.4 | 3.5 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.4 | 1.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.4 | 1.2 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.4 | 6.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.4 | 4.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.3 | 3.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.3 | 1.0 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.2 | 0.8 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.2 | 0.6 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.2 | 1.0 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.2 | 0.7 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.4 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.1 | 3.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.7 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 9.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 3.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.0 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.9 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.9 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 2.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.5 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 2.5 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.3 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 0.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 4.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.0 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.4 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 0.5 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 2.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 6.1 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 0.7 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 0.3 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.1 | 1.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 5.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.7 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 2.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.8 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 1.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 0.3 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 0.5 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 3.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 15.7 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.1 | 1.7 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.7 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.6 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 4.1 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.1 | 0.8 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.1 | 0.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 1.0 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0030586 | [methionine synthase] reductase activity(GO:0030586) |
| 0.0 | 2.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.6 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 6.9 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.3 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.7 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.4 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 1.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 10.3 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 1.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.2 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 0.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.9 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 1.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.4 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.4 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 1.7 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.3 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.4 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.0 | GO:0030251 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.0 | 0.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.3 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.3 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.2 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 2.0 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.2 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 10.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 3.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 0.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 6.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 1.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 1.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 2.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 10.0 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 3.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 0.9 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 2.7 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 3.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.3 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.7 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 1.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.1 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.2 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.6 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.3 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.2 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 1.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |