Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
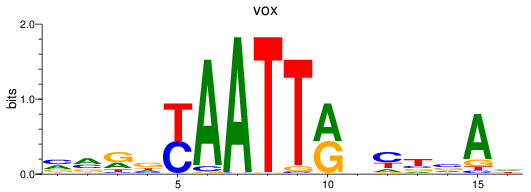
Results for vox
Z-value: 0.73
Transcription factors associated with vox
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
vox
|
ENSDARG00000099761 | ventral homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| vox | dr11_v1_chr13_-_50624173_50624173 | 0.97 | 3.5e-11 | Click! |
Activity profile of vox motif
Sorted Z-values of vox motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_29671586 | 2.63 |
ENSDART00000098336
|
spaca9
|
sperm acrosome associated 9 |
| chr23_+_45966436 | 2.30 |
ENSDART00000172160
|
CABZ01069338.1
|
|
| chr5_+_29671681 | 2.27 |
ENSDART00000043096
|
ak8
|
adenylate kinase 8 |
| chr7_-_40110140 | 2.03 |
ENSDART00000173469
|
si:ch73-174h16.5
|
si:ch73-174h16.5 |
| chr5_+_66353750 | 1.83 |
ENSDART00000143410
|
si:ch211-261c8.5
|
si:ch211-261c8.5 |
| chr20_-_14925281 | 1.69 |
ENSDART00000152641
|
dnm3a
|
dynamin 3a |
| chr1_-_17715493 | 1.63 |
ENSDART00000133027
|
si:dkey-256e7.8
|
si:dkey-256e7.8 |
| chr20_-_47188966 | 1.63 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr17_+_41992054 | 1.61 |
ENSDART00000182878
ENSDART00000111537 |
kiz
|
kizuna centrosomal protein |
| chr6_-_7720332 | 1.56 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr2_+_49799470 | 1.51 |
ENSDART00000146325
|
si:ch211-190k17.19
|
si:ch211-190k17.19 |
| chr5_+_66353589 | 1.51 |
ENSDART00000138246
|
si:ch211-261c8.5
|
si:ch211-261c8.5 |
| chr18_+_35173683 | 1.50 |
ENSDART00000192545
|
cfap45
|
cilia and flagella associated protein 45 |
| chr20_-_14924858 | 1.48 |
ENSDART00000047039
|
dnm3a
|
dynamin 3a |
| chr7_-_45999333 | 1.44 |
ENSDART00000158603
|
si:ch211-260e23.8
|
si:ch211-260e23.8 |
| chr25_+_19149241 | 1.43 |
ENSDART00000184982
ENSDART00000067324 |
mfge8b
|
milk fat globule-EGF factor 8 protein b |
| chr12_+_7445595 | 1.38 |
ENSDART00000103536
ENSDART00000152524 |
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr25_+_5035343 | 1.25 |
ENSDART00000011751
|
parvb
|
parvin, beta |
| chr1_+_22691256 | 1.24 |
ENSDART00000017413
|
zmynd10
|
zinc finger, MYND-type containing 10 |
| chr7_-_48667056 | 1.22 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr6_+_15762647 | 1.22 |
ENSDART00000127133
ENSDART00000128939 |
iqca1
|
IQ motif containing with AAA domain 1 |
| chr10_+_39091353 | 1.19 |
ENSDART00000125986
|
si:ch73-1a9.4
|
si:ch73-1a9.4 |
| chr6_+_30091811 | 1.11 |
ENSDART00000088403
|
meltf
|
melanotransferrin |
| chr16_+_54263921 | 1.10 |
ENSDART00000002856
|
drd2l
|
dopamine receptor D2 like |
| chr1_+_57311901 | 1.07 |
ENSDART00000149397
|
mycbpap
|
mycbp associated protein |
| chr5_+_7989210 | 1.04 |
ENSDART00000168071
|
gdnfb
|
glial cell derived neurotrophic factor b |
| chr5_+_19343880 | 1.03 |
ENSDART00000148130
|
acacb
|
acetyl-CoA carboxylase beta |
| chr21_+_6751405 | 0.98 |
ENSDART00000037265
ENSDART00000146371 |
olfm1b
|
olfactomedin 1b |
| chr14_+_30762131 | 0.96 |
ENSDART00000145039
|
si:ch211-145o7.3
|
si:ch211-145o7.3 |
| chr2_+_33747509 | 0.94 |
ENSDART00000134187
|
si:dkey-31m5.5
|
si:dkey-31m5.5 |
| chr25_+_31868268 | 0.93 |
ENSDART00000022325
|
parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr21_-_17296789 | 0.92 |
ENSDART00000192180
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr15_+_15856178 | 0.91 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr19_+_23932259 | 0.90 |
ENSDART00000139040
|
si:dkey-222b8.1
|
si:dkey-222b8.1 |
| chr10_-_5847655 | 0.87 |
ENSDART00000192773
|
ankrd55
|
ankyrin repeat domain 55 |
| chr10_+_35257651 | 0.85 |
ENSDART00000028940
|
styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr20_-_53435483 | 0.84 |
ENSDART00000135091
|
mettl24
|
methyltransferase like 24 |
| chr21_+_6751760 | 0.84 |
ENSDART00000135914
|
olfm1b
|
olfactomedin 1b |
| chr23_+_35713557 | 0.77 |
ENSDART00000123518
|
tuba1c
|
tubulin, alpha 1c |
| chr19_+_26718074 | 0.76 |
ENSDART00000134455
|
zgc:100906
|
zgc:100906 |
| chr7_+_6652967 | 0.71 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr13_+_8677166 | 0.69 |
ENSDART00000181016
ENSDART00000135028 |
prop1
|
PROP paired-like homeobox 1 |
| chr11_+_30057762 | 0.69 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr3_-_23643751 | 0.64 |
ENSDART00000078425
ENSDART00000140264 |
eve1
|
even-skipped-like1 |
| chr2_-_23411368 | 0.64 |
ENSDART00000159495
|
si:ch73-129a22.11
|
si:ch73-129a22.11 |
| chr2_-_48171441 | 0.63 |
ENSDART00000123040
|
pfkpb
|
phosphofructokinase, platelet b |
| chr5_-_64883082 | 0.62 |
ENSDART00000064983
ENSDART00000139066 |
krt1-c5
|
keratin, type 1, gene c5 |
| chr2_-_48171112 | 0.62 |
ENSDART00000156258
|
pfkpb
|
phosphofructokinase, platelet b |
| chr23_+_20110086 | 0.61 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr4_+_35553514 | 0.57 |
ENSDART00000182938
|
CR847906.1
|
|
| chr10_+_42690374 | 0.56 |
ENSDART00000123496
|
rhobtb2b
|
Rho-related BTB domain containing 2b |
| chr7_+_50822465 | 0.50 |
ENSDART00000011082
|
per1b
|
period circadian clock 1b |
| chr21_-_32097908 | 0.50 |
ENSDART00000147387
|
si:ch211-160j14.3
|
si:ch211-160j14.3 |
| chr25_-_4148719 | 0.49 |
ENSDART00000112880
ENSDART00000023278 |
fads2
|
fatty acid desaturase 2 |
| chr22_+_6905210 | 0.48 |
ENSDART00000167530
|
CABZ01065328.2
|
|
| chr6_-_30210378 | 0.45 |
ENSDART00000157359
ENSDART00000113924 |
lrrc7
|
leucine rich repeat containing 7 |
| chr4_+_5506952 | 0.45 |
ENSDART00000032857
ENSDART00000160222 |
mapk11
|
mitogen-activated protein kinase 11 |
| chr14_-_33105434 | 0.45 |
ENSDART00000163795
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr15_+_41027466 | 0.44 |
ENSDART00000075940
|
mtnr1ba
|
melatonin receptor type 1Ba |
| chr13_+_33688474 | 0.44 |
ENSDART00000161465
|
CABZ01087953.1
|
|
| chr11_+_16152316 | 0.43 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr3_-_15999501 | 0.42 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr8_+_17078692 | 0.41 |
ENSDART00000023206
|
plk2b
|
polo-like kinase 2b (Drosophila) |
| chr1_-_7582859 | 0.40 |
ENSDART00000110696
|
mxb
|
myxovirus (influenza) resistance B |
| chr10_-_40968095 | 0.39 |
ENSDART00000184104
|
npffr1l1
|
neuropeptide FF receptor 1 like 1 |
| chr18_+_48423973 | 0.37 |
ENSDART00000184233
ENSDART00000147074 |
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr5_+_66433287 | 0.37 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr8_+_52637507 | 0.35 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr16_-_7443388 | 0.35 |
ENSDART00000017445
|
prdm1a
|
PR domain containing 1a, with ZNF domain |
| chr25_-_18953322 | 0.34 |
ENSDART00000155927
|
si:ch211-68a17.7
|
si:ch211-68a17.7 |
| chr7_+_48667081 | 0.32 |
ENSDART00000083473
|
trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr11_-_29737088 | 0.32 |
ENSDART00000159828
|
si:ch211-218g23.6
|
si:ch211-218g23.6 |
| chr3_-_15444396 | 0.31 |
ENSDART00000104361
|
si:dkey-56d12.4
|
si:dkey-56d12.4 |
| chr15_-_15357178 | 0.29 |
ENSDART00000106120
|
ywhag2
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 |
| chr23_-_35082494 | 0.29 |
ENSDART00000189809
|
BX294434.1
|
|
| chr9_-_52962521 | 0.29 |
ENSDART00000170419
|
CU855885.1
|
|
| chr4_-_7212875 | 0.29 |
ENSDART00000161297
|
lrrn3b
|
leucine rich repeat neuronal 3b |
| chr15_+_5360407 | 0.29 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr6_-_35401282 | 0.28 |
ENSDART00000127612
|
rgs5a
|
regulator of G protein signaling 5a |
| chr1_-_49250490 | 0.28 |
ENSDART00000150386
|
si:ch73-6k14.2
|
si:ch73-6k14.2 |
| chr10_+_33744098 | 0.27 |
ENSDART00000147775
|
rxfp2a
|
relaxin/insulin-like family peptide receptor 2a |
| chr4_-_55459037 | 0.27 |
ENSDART00000172184
|
znf569l
|
zinc finger protein 569, like |
| chr2_-_22363460 | 0.26 |
ENSDART00000158486
|
selenof
|
selenoprotein F |
| chr10_+_21867307 | 0.26 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr17_+_33311784 | 0.25 |
ENSDART00000156229
|
si:ch211-132f19.7
|
si:ch211-132f19.7 |
| chr8_-_19246342 | 0.24 |
ENSDART00000147172
|
abhd17ab
|
abhydrolase domain containing 17Ab |
| chr19_+_7735157 | 0.24 |
ENSDART00000186717
|
tuft1b
|
tuftelin 1b |
| chr6_+_28051978 | 0.24 |
ENSDART00000143218
|
si:ch73-194h10.2
|
si:ch73-194h10.2 |
| chr22_-_17474583 | 0.24 |
ENSDART00000148027
|
si:ch211-197g15.8
|
si:ch211-197g15.8 |
| chr22_-_36530902 | 0.23 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr10_+_3049636 | 0.22 |
ENSDART00000081794
ENSDART00000183167 ENSDART00000191634 ENSDART00000183514 |
rasgrf2a
|
Ras protein-specific guanine nucleotide-releasing factor 2a |
| chr2_+_35240764 | 0.22 |
ENSDART00000015827
|
tnr
|
tenascin R (restrictin, janusin) |
| chr2_-_10896745 | 0.21 |
ENSDART00000114609
|
cdcp2
|
CUB domain containing protein 2 |
| chr11_+_2506516 | 0.20 |
ENSDART00000130886
ENSDART00000189767 |
NABP2
|
si:ch73-190f16.2 |
| chr18_+_8346920 | 0.19 |
ENSDART00000083421
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr6_-_39649504 | 0.18 |
ENSDART00000179960
ENSDART00000190951 |
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr15_+_3790556 | 0.18 |
ENSDART00000181467
|
RNF14 (1 of many)
|
ring finger protein 14 |
| chr10_+_37137464 | 0.18 |
ENSDART00000114909
|
cuedc1a
|
CUE domain containing 1a |
| chr3_+_34919810 | 0.16 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr6_-_39344259 | 0.15 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr22_-_17474781 | 0.15 |
ENSDART00000186817
|
si:ch211-197g15.8
|
si:ch211-197g15.8 |
| chr21_-_37790727 | 0.14 |
ENSDART00000162907
|
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr5_-_51484156 | 0.14 |
ENSDART00000162064
|
CR388055.1
|
|
| chr4_+_35594200 | 0.14 |
ENSDART00000193902
|
si:dkeyp-4c4.2
|
si:dkeyp-4c4.2 |
| chr11_+_38280454 | 0.13 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr21_+_15713097 | 0.12 |
ENSDART00000015841
|
gstt1b
|
glutathione S-transferase theta 1b |
| chr4_+_57580303 | 0.12 |
ENSDART00000166492
ENSDART00000103025 ENSDART00000170786 |
il17ra1b
il17ra2
|
interleukin 17 receptor A1b interleukin 17 receptor A2 |
| chr4_+_14727212 | 0.11 |
ENSDART00000158094
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr24_+_38301080 | 0.11 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr22_+_2198246 | 0.11 |
ENSDART00000165168
ENSDART00000157485 ENSDART00000168713 |
znf1157
znf1166
|
zinc finger protein 1157 zinc finger protein 1166 |
| chr16_-_25829779 | 0.10 |
ENSDART00000086301
|
irge4
|
immunity-related GTPase family, e4 |
| chr10_+_32104305 | 0.10 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr4_+_14727018 | 0.10 |
ENSDART00000124189
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr17_-_45525936 | 0.10 |
ENSDART00000084102
|
hhipl2
|
HHIP-like 2 |
| chr8_-_22274222 | 0.09 |
ENSDART00000131805
|
nphp4
|
nephronophthisis 4 |
| chr19_-_10971230 | 0.09 |
ENSDART00000166196
|
LO018584.1
|
|
| chr4_-_41269844 | 0.08 |
ENSDART00000186177
|
CR388165.2
|
|
| chr3_-_38783951 | 0.08 |
ENSDART00000155518
|
znf281a
|
zinc finger protein 281a |
| chr2_+_54798689 | 0.08 |
ENSDART00000183426
|
twsg1a
|
twisted gastrulation BMP signaling modulator 1a |
| chr11_+_24703108 | 0.07 |
ENSDART00000159173
|
gpr25
|
G protein-coupled receptor 25 |
| chr5_-_14373662 | 0.07 |
ENSDART00000183694
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr4_+_49322310 | 0.06 |
ENSDART00000184154
ENSDART00000167162 |
BX942819.1
|
|
| chr19_+_10592778 | 0.06 |
ENSDART00000135488
ENSDART00000151624 |
si:dkey-211g8.5
|
si:dkey-211g8.5 |
| chr5_+_56268436 | 0.05 |
ENSDART00000021159
|
lhx1b
|
LIM homeobox 1b |
| chr15_-_590787 | 0.05 |
ENSDART00000189367
|
si:ch73-144d13.5
|
si:ch73-144d13.5 |
| chr6_-_52348562 | 0.05 |
ENSDART00000142565
ENSDART00000145369 ENSDART00000016890 |
eif6
|
eukaryotic translation initiation factor 6 |
| chr22_-_7457247 | 0.05 |
ENSDART00000106081
|
BX511034.2
|
|
| chr2_-_33645411 | 0.05 |
ENSDART00000114663
|
b4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr20_+_37295006 | 0.04 |
ENSDART00000153137
|
cx23
|
connexin 23 |
| chr3_+_22334012 | 0.04 |
ENSDART00000193526
|
ifnphi3
|
interferon phi 3 |
| chr4_-_49119557 | 0.03 |
ENSDART00000150263
|
znf1036
|
zinc finger protein 1036 |
| chr5_+_28160503 | 0.03 |
ENSDART00000051516
|
tacr1a
|
tachykinin receptor 1a |
| chr2_+_35421285 | 0.03 |
ENSDART00000143787
|
tnr
|
tenascin R (restrictin, janusin) |
| chr11_-_7380674 | 0.03 |
ENSDART00000014979
ENSDART00000103418 |
vtg3
|
vitellogenin 3, phosvitinless |
| chr12_-_7157527 | 0.03 |
ENSDART00000152274
|
si:dkey-187i8.2
|
si:dkey-187i8.2 |
| chr15_-_2493771 | 0.03 |
ENSDART00000184906
|
neu4
|
sialidase 4 |
| chr15_-_2497568 | 0.02 |
ENSDART00000080398
|
neu4
|
sialidase 4 |
| chr20_+_26966725 | 0.01 |
ENSDART00000029781
|
ahsa1a
|
AHA1, activator of heat shock protein ATPase homolog 1a |
| chr23_-_7674902 | 0.00 |
ENSDART00000185612
ENSDART00000180524 |
plagl2
|
pleiomorphic adenoma gene-like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of vox
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.3 | 1.0 | GO:0061213 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.3 | 1.0 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.2 | 0.8 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.2 | 3.6 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.2 | 1.2 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 1.2 | GO:0006735 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 1.1 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 1.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.3 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.6 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.1 | 1.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.4 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 2.7 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 | 1.1 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.5 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.9 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.7 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 0.3 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.9 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.5 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 1.4 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.3 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.2 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.0 | GO:0042256 | mature ribosome assembly(GO:0042256) assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.6 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.4 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.1 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 1.6 | GO:0007051 | spindle organization(GO:0007051) |
| 0.0 | 0.6 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 0.4 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.2 | GO:0009437 | carnitine metabolic process(GO:0009437) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 3.6 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 1.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.9 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 1.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.1 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.1 | GO:0055037 | recycling endosome(GO:0055037) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.3 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.3 | 1.0 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.3 | 1.0 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 1.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 1.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.7 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 1.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.4 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 1.6 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.9 | GO:0033549 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.9 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.2 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.3 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |