Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
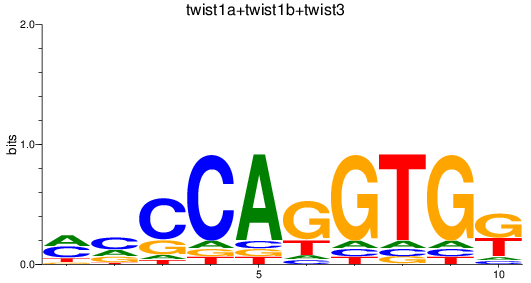
Results for twist1a+twist1b+twist3
Z-value: 0.29
Transcription factors associated with twist1a+twist1b+twist3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
twist3
|
ENSDARG00000019646 | twist3 |
|
twist1a
|
ENSDARG00000030402 | twist family bHLH transcription factor 1a |
|
twist1b
|
ENSDARG00000076010 | twist family bHLH transcription factor 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| twist1b | dr11_v1_chr16_+_19732543_19732543 | -0.95 | 1.0e-09 | Click! |
| twist1a | dr11_v1_chr19_-_2231146_2231188 | 0.51 | 3.1e-02 | Click! |
| twist3 | dr11_v1_chr23_+_2669_2669 | -0.17 | 4.9e-01 | Click! |
Activity profile of twist1a+twist1b+twist3 motif
Sorted Z-values of twist1a+twist1b+twist3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_44574059 | 1.20 |
ENSDART00000123007
|
si:ch73-160p18.3
|
si:ch73-160p18.3 |
| chr21_-_43666420 | 1.07 |
ENSDART00000139008
ENSDART00000183996 ENSDART00000183395 |
si:dkey-229d11.3
si:dkey-229d11.5
|
si:dkey-229d11.3 si:dkey-229d11.5 |
| chr17_+_1360192 | 0.54 |
ENSDART00000184561
|
siva1
|
SIVA1, apoptosis-inducing factor |
| chr5_+_6670945 | 0.52 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr10_+_1052591 | 0.43 |
ENSDART00000123405
|
unc5c
|
unc-5 netrin receptor C |
| chr5_-_30080332 | 0.39 |
ENSDART00000140049
|
bco2a
|
beta-carotene oxygenase 2a |
| chr21_-_35325466 | 0.38 |
ENSDART00000134780
ENSDART00000145930 ENSDART00000076715 ENSDART00000065341 ENSDART00000162189 |
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr3_+_58472305 | 0.37 |
ENSDART00000154122
|
si:ch211-165g14.1
|
si:ch211-165g14.1 |
| chr5_-_23696926 | 0.34 |
ENSDART00000021462
|
rnf128a
|
ring finger protein 128a |
| chr14_-_31087830 | 0.34 |
ENSDART00000002250
|
hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr16_-_19568795 | 0.32 |
ENSDART00000185141
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr20_-_44575103 | 0.31 |
ENSDART00000192573
|
ubxn2a
|
UBX domain protein 2A |
| chr6_-_10728921 | 0.31 |
ENSDART00000151484
|
sp3b
|
Sp3b transcription factor |
| chr18_+_45666489 | 0.29 |
ENSDART00000180147
ENSDART00000151351 |
prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr11_-_34232906 | 0.29 |
ENSDART00000162150
|
lsg1
|
large 60S subunit nuclear export GTPase 1 |
| chr5_-_32505109 | 0.29 |
ENSDART00000188219
|
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr6_-_55423220 | 0.28 |
ENSDART00000158929
|
ctsa
|
cathepsin A |
| chr16_-_31791165 | 0.28 |
ENSDART00000148389
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr6_-_10728057 | 0.27 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr23_+_2914577 | 0.25 |
ENSDART00000184897
|
DHX35
|
zgc:158828 |
| chr4_+_26496489 | 0.23 |
ENSDART00000160652
|
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr3_+_16663373 | 0.22 |
ENSDART00000100961
|
zgc:55558
|
zgc:55558 |
| chr1_+_45925150 | 0.20 |
ENSDART00000074689
|
eif5b
|
eukaryotic translation initiation factor 5B |
| chr1_+_45925365 | 0.19 |
ENSDART00000144245
|
eif5b
|
eukaryotic translation initiation factor 5B |
| chr17_-_40876680 | 0.19 |
ENSDART00000127200
|
supt7l
|
SPT7-like STAGA complex gamma subunit |
| chr5_-_11809404 | 0.18 |
ENSDART00000132564
|
nf2a
|
neurofibromin 2a (merlin) |
| chr7_-_9674073 | 0.17 |
ENSDART00000187902
|
lrrk1
|
leucine-rich repeat kinase 1 |
| chr18_-_35842554 | 0.15 |
ENSDART00000088488
|
opa3
|
optic atrophy 3 |
| chr1_-_55058795 | 0.14 |
ENSDART00000187293
|
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr8_+_20140321 | 0.14 |
ENSDART00000012120
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr3_+_59411956 | 0.14 |
ENSDART00000166982
|
sec14l1
|
SEC14-like lipid binding 1 |
| chr16_-_21047483 | 0.12 |
ENSDART00000136235
|
cbx3b
|
chromobox homolog 3b |
| chr6_+_42693114 | 0.11 |
ENSDART00000154353
|
si:ch211-207d10.2
|
si:ch211-207d10.2 |
| chr20_-_22193190 | 0.11 |
ENSDART00000047624
|
tmem165
|
transmembrane protein 165 |
| chr20_+_36623807 | 0.10 |
ENSDART00000149171
ENSDART00000062895 |
srp9
|
signal recognition particle 9 |
| chr5_-_15321451 | 0.10 |
ENSDART00000139203
|
txnrd2.1
|
thioredoxin reductase 2, tandem duplicate 1 |
| chr13_-_9119867 | 0.09 |
ENSDART00000137255
|
si:dkey-112g5.15
|
si:dkey-112g5.15 |
| chr3_+_49021079 | 0.09 |
ENSDART00000162012
|
zgc:163083
|
zgc:163083 |
| chr24_+_39660124 | 0.08 |
ENSDART00000066500
|
stub1
|
STIP1 homology and U-Box containing protein 1 |
| chr2_+_38002717 | 0.06 |
ENSDART00000139564
|
dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr6_+_42693288 | 0.05 |
ENSDART00000155010
|
si:ch211-207d10.2
|
si:ch211-207d10.2 |
| chr22_-_26251563 | 0.05 |
ENSDART00000060888
ENSDART00000142821 |
ccdc130
|
coiled-coil domain containing 130 |
| chr12_-_44180132 | 0.05 |
ENSDART00000165998
|
si:ch73-329n5.1
|
si:ch73-329n5.1 |
| chr15_-_46736432 | 0.04 |
ENSDART00000124567
|
ilkap
|
integrin-linked kinase-associated serine/threonine phosphatase |
| chr11_-_40101246 | 0.04 |
ENSDART00000161083
|
tnfrsf9b
|
tumor necrosis factor receptor superfamily, member 9b |
| chr15_-_766015 | 0.04 |
ENSDART00000190648
|
si:dkey-7i4.15
|
si:dkey-7i4.15 |
| chr18_-_21859019 | 0.03 |
ENSDART00000100885
|
nrn1la
|
neuritin 1-like a |
| chr7_+_17485782 | 0.03 |
ENSDART00000098083
|
CU179759.1
|
Danio rerio novel immune-type receptor 1d (nitr1d), mRNA. |
| chr5_-_65159258 | 0.03 |
ENSDART00000160429
|
sh2d3cb
|
SH2 domain containing 3Cb |
| chr4_-_36476889 | 0.03 |
ENSDART00000163956
|
si:ch211-263l8.1
|
si:ch211-263l8.1 |
| chr16_-_21047872 | 0.02 |
ENSDART00000131582
|
cbx3b
|
chromobox homolog 3b |
| chr4_-_31700186 | 0.01 |
ENSDART00000183986
ENSDART00000180890 |
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr4_+_33830944 | 0.01 |
ENSDART00000170238
|
znf1026
|
zinc finger protein 1026 |
| chr8_+_247163 | 0.01 |
ENSDART00000122378
|
cep120
|
centrosomal protein 120 |
| chr9_-_327901 | 0.00 |
ENSDART00000159956
|
ndufa4l2a
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2a |
| chr4_+_1530287 | 0.00 |
ENSDART00000067446
|
slc38a4
|
solute carrier family 38, member 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of twist1a+twist1b+twist3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0050787 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.1 | 0.3 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.4 | GO:0042214 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.4 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.5 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.1 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.0 | 0.3 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.3 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.1 | 0.3 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.4 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.5 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 0.1 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |