Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
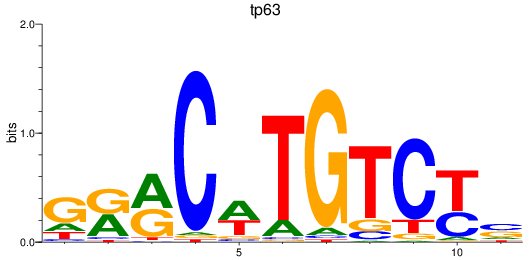
Results for tp63
Z-value: 0.47
Transcription factors associated with tp63
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tp63
|
ENSDARG00000044356 | tumor protein p63 |
|
tp63
|
ENSDARG00000113627 | tumor protein p63 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tp63 | dr11_v1_chr6_+_28796773_28796773 | 0.42 | 8.1e-02 | Click! |
Activity profile of tp63 motif
Sorted Z-values of tp63 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_21672700 | 1.66 |
ENSDART00000187043
|
si:dkey-40g16.5
|
si:dkey-40g16.5 |
| chr12_+_45200744 | 1.48 |
ENSDART00000098932
|
wbp2
|
WW domain binding protein 2 |
| chr15_+_44053244 | 1.43 |
ENSDART00000059550
|
lrrc51
|
leucine rich repeat containing 51 |
| chr2_-_32501501 | 1.41 |
ENSDART00000181309
|
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr6_+_3280939 | 0.90 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr10_+_39091353 | 0.80 |
ENSDART00000125986
|
si:ch73-1a9.4
|
si:ch73-1a9.4 |
| chr17_+_53250802 | 0.72 |
ENSDART00000143819
|
VASH1
|
vasohibin 1 |
| chr20_-_1314537 | 0.70 |
ENSDART00000144288
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr22_-_24984733 | 0.64 |
ENSDART00000142147
ENSDART00000187284 |
dnal4b
|
dynein, axonemal, light chain 4b |
| chr25_+_34641536 | 0.63 |
ENSDART00000167033
|
CABZ01079011.1
|
|
| chr2_-_26563978 | 0.63 |
ENSDART00000150016
|
glis1a
|
GLIS family zinc finger 1a |
| chr10_+_25355308 | 0.61 |
ENSDART00000100415
|
map3k7cl
|
map3k7 C-terminal like |
| chr1_+_52735484 | 0.61 |
ENSDART00000182076
|
CABZ01021532.1
|
|
| chr20_+_36629173 | 0.60 |
ENSDART00000161241
|
ephx1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr7_-_50914526 | 0.57 |
ENSDART00000160398
|
ankrd46b
|
ankyrin repeat domain 46b |
| chr23_+_44307996 | 0.55 |
ENSDART00000042430
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr20_-_1314355 | 0.53 |
ENSDART00000152436
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr14_-_9522364 | 0.51 |
ENSDART00000054689
|
atoh8
|
atonal bHLH transcription factor 8 |
| chr17_-_7861219 | 0.48 |
ENSDART00000148604
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr4_-_52165969 | 0.45 |
ENSDART00000171130
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr14_+_24241241 | 0.45 |
ENSDART00000022377
|
nkx2.5
|
NK2 homeobox 5 |
| chr24_+_35564668 | 0.42 |
ENSDART00000122734
|
cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr25_+_4787431 | 0.40 |
ENSDART00000170640
|
myo5c
|
myosin VC |
| chr10_+_5135842 | 0.37 |
ENSDART00000132627
ENSDART00000162434 |
zgc:113274
|
zgc:113274 |
| chr7_+_8324506 | 0.36 |
ENSDART00000168110
|
si:dkey-185m8.2
|
si:dkey-185m8.2 |
| chr17_-_43466317 | 0.36 |
ENSDART00000155313
|
hspa4l
|
heat shock protein 4 like |
| chr17_-_14613711 | 0.35 |
ENSDART00000157345
|
sdsl
|
serine dehydratase-like |
| chr23_-_1056808 | 0.33 |
ENSDART00000081961
|
zgc:113423
|
zgc:113423 |
| chr11_-_11518469 | 0.32 |
ENSDART00000104254
|
krt15
|
keratin 15 |
| chr17_-_22324727 | 0.28 |
ENSDART00000160341
|
CU104709.1
|
|
| chr8_+_2487883 | 0.28 |
ENSDART00000101841
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr9_+_41103127 | 0.27 |
ENSDART00000000250
|
slc40a1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr25_+_4787607 | 0.27 |
ENSDART00000159422
|
myo5c
|
myosin VC |
| chr13_-_12581388 | 0.26 |
ENSDART00000079655
|
enpep
|
glutamyl aminopeptidase |
| chr18_+_17611627 | 0.24 |
ENSDART00000046891
|
cetp
|
cholesteryl ester transfer protein, plasma |
| chr8_-_4694458 | 0.23 |
ENSDART00000019828
|
zgc:63587
|
zgc:63587 |
| chr6_+_54538948 | 0.22 |
ENSDART00000149270
|
tulp1b
|
tubby like protein 1b |
| chr25_+_31912153 | 0.22 |
ENSDART00000153968
|
apba2a
|
amyloid beta (A4) precursor protein-binding, family A, member 2a |
| chr17_-_32863250 | 0.21 |
ENSDART00000167292
|
prox1a
|
prospero homeobox 1a |
| chr7_-_60156409 | 0.19 |
ENSDART00000006802
|
cct7
|
chaperonin containing TCP1, subunit 7 (eta) |
| chr25_+_22107643 | 0.18 |
ENSDART00000089680
|
sigirr
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr7_+_69653981 | 0.18 |
ENSDART00000090165
|
frem1a
|
Fras1 related extracellular matrix 1a |
| chr25_+_17871089 | 0.16 |
ENSDART00000133725
|
btbd10a
|
BTB (POZ) domain containing 10a |
| chr15_-_29387446 | 0.15 |
ENSDART00000145976
ENSDART00000035096 |
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr20_-_25436389 | 0.14 |
ENSDART00000153266
|
itsn2a
|
intersectin 2a |
| chr2_+_36691771 | 0.14 |
ENSDART00000131978
|
ptx3b
|
pentraxin 3, long b |
| chr16_+_20904754 | 0.14 |
ENSDART00000006043
|
hoxa11b
|
homeobox A11b |
| chr20_-_27325258 | 0.13 |
ENSDART00000152917
|
asb2a.1
|
ankyrin repeat and SOCS box containing 2a, tandem duplicate 1 |
| chr7_-_56606752 | 0.12 |
ENSDART00000138714
|
sult5a1
|
sulfotransferase family 5A, member 1 |
| chr10_-_10607118 | 0.11 |
ENSDART00000101089
|
dbh
|
dopamine beta-hydroxylase (dopamine beta-monooxygenase) |
| chr23_-_19827411 | 0.11 |
ENSDART00000187964
|
haus7
|
HAUS augmin-like complex, subunit 7 |
| chr18_-_17399291 | 0.10 |
ENSDART00000192075
ENSDART00000060949 ENSDART00000188506 |
zfpm1
|
zinc finger protein, FOG family member 1 |
| chr13_-_41908583 | 0.09 |
ENSDART00000136515
|
ipmka
|
inositol polyphosphate multikinase a |
| chr15_+_14378829 | 0.08 |
ENSDART00000160145
|
si:ch211-11n16.2
|
si:ch211-11n16.2 |
| chr5_+_38913621 | 0.07 |
ENSDART00000137112
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr13_+_25422009 | 0.06 |
ENSDART00000057686
|
calhm2
|
calcium homeostasis modulator 2 |
| chr22_+_21305682 | 0.05 |
ENSDART00000023521
|
odf3l2
|
outer dense fiber of sperm tails 3-like 2 |
| chr22_+_569565 | 0.05 |
ENSDART00000037069
|
usp49
|
ubiquitin specific peptidase 49 |
| chr16_+_48682309 | 0.04 |
ENSDART00000183957
|
tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr23_-_17470146 | 0.03 |
ENSDART00000002398
|
trim101
|
tripartite motif containing 101 |
| chr18_-_33080454 | 0.03 |
ENSDART00000191907
|
v2ra18
|
vomeronasal 2 receptor, a18 |
| chr1_+_36651059 | 0.03 |
ENSDART00000187475
|
ednraa
|
endothelin receptor type Aa |
| chr7_+_22702225 | 0.02 |
ENSDART00000173672
|
si:dkey-165a24.9
|
si:dkey-165a24.9 |
| chr17_+_43623598 | 0.02 |
ENSDART00000154138
|
znf365
|
zinc finger protein 365 |
| chr13_+_25421531 | 0.01 |
ENSDART00000158093
|
calhm2
|
calcium homeostasis modulator 2 |
| chr17_+_38724473 | 0.01 |
ENSDART00000075792
|
GPR68
|
G protein-coupled receptor 68 |
| chr17_+_45607580 | 0.01 |
ENSDART00000122583
|
si:ch211-202f3.4
|
si:ch211-202f3.4 |
| chr17_+_30450163 | 0.00 |
ENSDART00000104257
|
lpin1
|
lipin 1 |
| chr8_-_38506339 | 0.00 |
ENSDART00000054558
ENSDART00000141623 |
foxb2
|
forkhead box B2 |
| chr21_-_45613564 | 0.00 |
ENSDART00000160324
|
LO018363.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of tp63
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.2 | 0.5 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.2 | 0.9 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.4 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.1 | 0.4 | GO:0009097 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.1 | 0.2 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.1 | 0.3 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 1.5 | GO:0045815 | positive regulation of gene expression, epigenetic(GO:0045815) |
| 0.0 | 0.1 | GO:0042421 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.0 | 0.1 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.5 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.6 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.7 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.2 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.0 | GO:0019884 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.6 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 0.6 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.1 | 0.4 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.9 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.3 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.9 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 1.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.3 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |