Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for tlx1
Z-value: 0.44
Transcription factors associated with tlx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
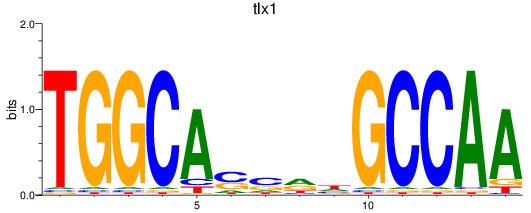
tlx1
|
ENSDARG00000003965 | T cell leukemia homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tlx1 | dr11_v1_chr13_-_28272299_28272299 | 0.13 | 5.9e-01 | Click! |
Activity profile of tlx1 motif
Sorted Z-values of tlx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_17232372 | 1.45 |
ENSDART00000135679
|
rab36
|
RAB36, member RAS oncogene family |
| chr19_+_9004256 | 1.42 |
ENSDART00000186401
ENSDART00000091443 |
si:ch211-81a5.1
|
si:ch211-81a5.1 |
| chr25_+_13158549 | 0.94 |
ENSDART00000167804
|
si:dkeyp-50b9.1
|
si:dkeyp-50b9.1 |
| chr13_+_37040789 | 0.87 |
ENSDART00000063412
|
esr2b
|
estrogen receptor 2b |
| chr6_+_26346686 | 0.80 |
ENSDART00000154183
|
dzip1l
|
DAZ interacting zinc finger protein 1-like |
| chr7_-_35126374 | 0.77 |
ENSDART00000141211
|
hsd11b2
|
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr22_+_22888 | 0.67 |
ENSDART00000082471
|
mfap2
|
microfibril associated protein 2 |
| chr3_+_57038033 | 0.50 |
ENSDART00000162930
|
bahcc1a
|
BAH domain and coiled-coil containing 1a |
| chr2_-_17827983 | 0.50 |
ENSDART00000166518
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr13_+_15682803 | 0.49 |
ENSDART00000188063
|
CR931980.1
|
|
| chr7_+_14291323 | 0.47 |
ENSDART00000053521
|
rhcga
|
Rh family, C glycoprotein a |
| chr2_-_21618629 | 0.46 |
ENSDART00000027587
|
ralab
|
v-ral simian leukemia viral oncogene homolog Ab (ras related) |
| chr4_-_16406737 | 0.45 |
ENSDART00000013085
|
dcn
|
decorin |
| chr16_+_23397785 | 0.44 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr20_+_16750177 | 0.44 |
ENSDART00000185357
|
calm1b
|
calmodulin 1b |
| chr4_+_61171310 | 0.43 |
ENSDART00000141738
|
si:dkey-9p20.18
|
si:dkey-9p20.18 |
| chr25_-_4146947 | 0.42 |
ENSDART00000129268
|
fads2
|
fatty acid desaturase 2 |
| chr2_-_2937225 | 0.42 |
ENSDART00000168132
|
si:ch1073-82l19.1
|
si:ch1073-82l19.1 |
| chr1_+_49568335 | 0.41 |
ENSDART00000142957
|
col17a1a
|
collagen, type XVII, alpha 1a |
| chr9_-_22831836 | 0.40 |
ENSDART00000142585
|
neb
|
nebulin |
| chr10_+_8091144 | 0.39 |
ENSDART00000143848
ENSDART00000075485 |
sub1a
|
SUB1 homolog, transcriptional regulator a |
| chr24_-_20369604 | 0.38 |
ENSDART00000143174
|
dlec1
|
deleted in lung and esophageal cancer 1 |
| chr19_-_30404096 | 0.37 |
ENSDART00000103475
|
agr2
|
anterior gradient 2 |
| chr17_-_29771639 | 0.34 |
ENSDART00000086201
|
ush2a
|
Usher syndrome 2A (autosomal recessive, mild) |
| chr4_-_38033800 | 0.33 |
ENSDART00000159662
|
si:dkeyp-82b4.4
|
si:dkeyp-82b4.4 |
| chr20_-_2134620 | 0.31 |
ENSDART00000064375
|
tmem244
|
transmembrane protein 244 |
| chr10_-_15672862 | 0.31 |
ENSDART00000109231
|
mamdc2b
|
MAM domain containing 2b |
| chr18_-_39473055 | 0.31 |
ENSDART00000122930
|
scg3
|
secretogranin III |
| chr2_+_55982940 | 0.31 |
ENSDART00000097753
ENSDART00000097751 |
nmrk2
|
nicotinamide riboside kinase 2 |
| chr23_-_20100436 | 0.30 |
ENSDART00000143445
|
si:dkey-32e6.6
|
si:dkey-32e6.6 |
| chr10_+_8091690 | 0.29 |
ENSDART00000189915
|
sub1a
|
SUB1 homolog, transcriptional regulator a |
| chr24_-_21819010 | 0.28 |
ENSDART00000091096
|
CR352265.1
|
|
| chr2_-_8017579 | 0.28 |
ENSDART00000040209
|
ephb3a
|
eph receptor B3a |
| chr3_-_15081874 | 0.28 |
ENSDART00000192532
|
nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr23_-_45504991 | 0.27 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr7_-_22132265 | 0.27 |
ENSDART00000125284
ENSDART00000112978 |
nlgn2a
|
neuroligin 2a |
| chr5_-_67629263 | 0.26 |
ENSDART00000133753
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr4_-_64709908 | 0.26 |
ENSDART00000161032
|
si:dkey-9i5.2
|
si:dkey-9i5.2 |
| chr24_-_20369760 | 0.26 |
ENSDART00000185585
|
dlec1
|
deleted in lung and esophageal cancer 1 |
| chr20_+_10545514 | 0.26 |
ENSDART00000153667
|
serpina1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr19_-_30403922 | 0.26 |
ENSDART00000181841
|
agr2
|
anterior gradient 2 |
| chr20_+_10539293 | 0.25 |
ENSDART00000182265
|
serpina1l
|
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like |
| chr2_-_1364678 | 0.25 |
ENSDART00000011919
ENSDART00000164674 |
ss18
|
synovial sarcoma translocation, chromosome 18 (H. sapiens) |
| chr22_-_22416337 | 0.24 |
ENSDART00000142947
ENSDART00000089569 |
camsap2a
|
calmodulin regulated spectrin-associated protein family, member 2a |
| chr17_-_7496341 | 0.22 |
ENSDART00000186907
|
si:ch211-278p9.3
|
si:ch211-278p9.3 |
| chr7_+_57109214 | 0.21 |
ENSDART00000135068
ENSDART00000098412 |
enosf1
|
enolase superfamily member 1 |
| chr12_-_36521947 | 0.21 |
ENSDART00000152946
|
unc13d
|
unc-13 homolog D (C. elegans) |
| chr17_+_52822831 | 0.20 |
ENSDART00000193368
|
meis2a
|
Meis homeobox 2a |
| chr17_+_37645633 | 0.19 |
ENSDART00000085500
|
tmem229b
|
transmembrane protein 229B |
| chr13_-_8888334 | 0.19 |
ENSDART00000059881
|
plekhh2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 |
| chr22_+_39058269 | 0.19 |
ENSDART00000113362
|
ip6k1
|
inositol hexakisphosphate kinase 1 |
| chr4_-_6416414 | 0.18 |
ENSDART00000191136
|
MDFIC
|
si:ch73-156e19.1 |
| chr12_+_35654749 | 0.18 |
ENSDART00000169889
ENSDART00000167873 |
baiap2b
|
BAI1-associated protein 2b |
| chr21_+_5257018 | 0.17 |
ENSDART00000183100
ENSDART00000191525 |
loxhd1a
|
lipoxygenase homology domains 1a |
| chr23_+_21278948 | 0.17 |
ENSDART00000156701
ENSDART00000033970 |
ubr4
|
ubiquitin protein ligase E3 component n-recognin 4 |
| chr7_+_57108823 | 0.16 |
ENSDART00000184943
ENSDART00000055956 |
enosf1
|
enolase superfamily member 1 |
| chr6_+_39184236 | 0.16 |
ENSDART00000156187
|
tac3b
|
tachykinin 3b |
| chr12_+_22607761 | 0.16 |
ENSDART00000153112
|
si:dkey-219e21.2
|
si:dkey-219e21.2 |
| chr4_-_6416155 | 0.15 |
ENSDART00000110535
|
MDFIC
|
si:ch73-156e19.1 |
| chr2_+_30969029 | 0.15 |
ENSDART00000085242
|
lpin2
|
lipin 2 |
| chr7_-_28647959 | 0.15 |
ENSDART00000150148
|
slc7a6
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6 |
| chr15_+_22722684 | 0.15 |
ENSDART00000156760
|
grik4
|
glutamate receptor, ionotropic, kainate 4 |
| chr13_-_32577386 | 0.14 |
ENSDART00000016535
|
kcns3a
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3a |
| chr20_-_20410029 | 0.13 |
ENSDART00000192177
ENSDART00000063483 |
prkchb
|
protein kinase C, eta, b |
| chr1_-_56363108 | 0.13 |
ENSDART00000021878
|
gcgrb
|
glucagon receptor b |
| chr8_-_34052019 | 0.13 |
ENSDART00000040126
ENSDART00000159208 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr16_+_21426524 | 0.13 |
ENSDART00000182869
|
gsdmeb
|
gasdermin Eb |
| chr9_-_24242592 | 0.13 |
ENSDART00000039399
|
cavin2a
|
caveolae associated protein 2a |
| chr8_-_53108207 | 0.13 |
ENSDART00000111023
|
b3galt4
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 |
| chr6_-_35401282 | 0.13 |
ENSDART00000127612
|
rgs5a
|
regulator of G protein signaling 5a |
| chr19_+_30388186 | 0.13 |
ENSDART00000103474
|
tspan13b
|
tetraspanin 13b |
| chr8_+_44722140 | 0.12 |
ENSDART00000163381
|
elmod3
|
ELMO/CED-12 domain containing 3 |
| chr1_-_22757145 | 0.12 |
ENSDART00000134719
|
prom1b
|
prominin 1 b |
| chr8_-_31053872 | 0.12 |
ENSDART00000109885
|
snrnp200
|
small nuclear ribonucleoprotein 200 (U5) |
| chr3_-_37681824 | 0.11 |
ENSDART00000185858
|
gpatch8
|
G patch domain containing 8 |
| chr25_-_30394746 | 0.11 |
ENSDART00000185346
|
si:ch211-93f2.1
|
si:ch211-93f2.1 |
| chr16_+_16849220 | 0.11 |
ENSDART00000047409
ENSDART00000142155 |
myh14
|
myosin, heavy chain 14, non-muscle |
| chr20_+_2134816 | 0.11 |
ENSDART00000039249
|
l3mbtl3
|
l(3)mbt-like 3 (Drosophila) |
| chr9_-_34296406 | 0.10 |
ENSDART00000125451
|
ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr22_-_11438627 | 0.10 |
ENSDART00000007649
|
mid1ip1b
|
MID1 interacting protein 1b |
| chr1_+_35194454 | 0.10 |
ENSDART00000165389
|
sc:d189
|
sc:d189 |
| chr8_+_53423408 | 0.10 |
ENSDART00000164792
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr14_-_32503363 | 0.10 |
ENSDART00000034883
|
mcf2a
|
MCF.2 cell line derived transforming sequence a |
| chr17_+_52822422 | 0.10 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr16_+_14029283 | 0.09 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr4_+_31450683 | 0.09 |
ENSDART00000169303
|
znf996
|
zinc finger protein 996 |
| chr18_-_2433011 | 0.09 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr18_-_43880020 | 0.08 |
ENSDART00000185638
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr15_-_14469704 | 0.08 |
ENSDART00000185077
|
numbl
|
numb homolog (Drosophila)-like |
| chr7_-_24472991 | 0.08 |
ENSDART00000121684
|
nat8l
|
N-acetyltransferase 8-like |
| chr3_-_34599662 | 0.08 |
ENSDART00000055259
|
nmrk1
|
nicotinamide riboside kinase 1 |
| chr13_+_4888351 | 0.08 |
ENSDART00000145940
|
micu1
|
mitochondrial calcium uptake 1 |
| chr11_-_270210 | 0.08 |
ENSDART00000005217
ENSDART00000172779 |
alas1
|
aminolevulinate, delta-, synthase 1 |
| chr17_+_5976683 | 0.07 |
ENSDART00000110276
|
zgc:194275
|
zgc:194275 |
| chr7_+_31051603 | 0.07 |
ENSDART00000108721
|
tjp1a
|
tight junction protein 1a |
| chr17_-_22048233 | 0.07 |
ENSDART00000155203
|
ttbk1b
|
tau tubulin kinase 1b |
| chr4_+_47385355 | 0.06 |
ENSDART00000163413
|
znf1122
|
zinc finger protein 1122 |
| chr18_+_40462445 | 0.06 |
ENSDART00000087645
|
ugt5c2
|
UDP glucuronosyltransferase 5 family, polypeptide C2 |
| chr12_-_2518178 | 0.06 |
ENSDART00000128677
ENSDART00000086510 |
mapk8b
|
mitogen-activated protein kinase 8b |
| chr2_-_59285085 | 0.06 |
ENSDART00000131880
|
ftr34
|
finTRIM family, member 34 |
| chr11_+_24251141 | 0.06 |
ENSDART00000182684
|
pnp4a
|
purine nucleoside phosphorylase 4a |
| chr6_+_28141135 | 0.06 |
ENSDART00000113405
ENSDART00000179795 |
inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr3_-_19561058 | 0.05 |
ENSDART00000079323
|
zgc:163079
|
zgc:163079 |
| chr6_-_13114406 | 0.05 |
ENSDART00000188015
|
zgc:194469
|
zgc:194469 |
| chr20_+_25581627 | 0.05 |
ENSDART00000030229
|
cyp2p9
|
cytochrome P450, family 2, subfamily P, polypeptide 9 |
| chr24_+_25692802 | 0.05 |
ENSDART00000190493
|
camk2n2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chr20_-_33976305 | 0.05 |
ENSDART00000111022
|
sele
|
selectin E |
| chr17_+_30546579 | 0.05 |
ENSDART00000154385
|
nhsl1a
|
NHS-like 1a |
| chr5_+_35502290 | 0.05 |
ENSDART00000191222
|
CR762390.1
|
|
| chr13_-_15865335 | 0.05 |
ENSDART00000135186
|
si:ch211-57f7.7
|
si:ch211-57f7.7 |
| chr20_+_34320635 | 0.05 |
ENSDART00000153207
|
ivns1abpa
|
influenza virus NS1A binding protein a |
| chr22_+_11857356 | 0.05 |
ENSDART00000179540
|
mras
|
muscle RAS oncogene homolog |
| chr13_+_18371208 | 0.04 |
ENSDART00000138172
|
ccar1
|
cell division cycle and apoptosis regulator 1 |
| chr12_-_37941733 | 0.04 |
ENSDART00000130167
|
CABZ01046949.1
|
|
| chr8_-_4031121 | 0.04 |
ENSDART00000169474
ENSDART00000163754 |
mtmr3
|
myotubularin related protein 3 |
| chr17_-_31308658 | 0.04 |
ENSDART00000124505
|
bahd1
|
bromo adjacent homology domain containing 1 |
| chr16_-_9453591 | 0.03 |
ENSDART00000126154
|
prpf3
|
PRP3 pre-mRNA processing factor 3 homolog (yeast) |
| chr4_-_51460013 | 0.03 |
ENSDART00000193382
|
CR628395.1
|
|
| chr9_+_35860975 | 0.03 |
ENSDART00000134447
|
rcan1a
|
regulator of calcineurin 1a |
| chr13_+_28761998 | 0.03 |
ENSDART00000146581
|
prom2
|
prominin 2 |
| chr7_+_23292133 | 0.03 |
ENSDART00000134489
|
htr2cl1
|
5-hydroxytryptamine (serotonin) receptor 2C, G protein-coupled-like 1 |
| chr17_+_38262408 | 0.03 |
ENSDART00000017493
|
nkx2.1
|
NK2 homeobox 1 |
| chr4_-_46648979 | 0.02 |
ENSDART00000162101
|
BX927193.2
|
|
| chr8_-_39822917 | 0.02 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr21_-_2707768 | 0.02 |
ENSDART00000165384
|
CABZ01101739.1
|
|
| chr18_+_41560822 | 0.02 |
ENSDART00000158503
|
baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr3_+_33918510 | 0.02 |
ENSDART00000055248
|
alkbh7
|
alkB homolog 7 |
| chr21_+_39336285 | 0.02 |
ENSDART00000139677
|
si:ch211-274p24.4
|
si:ch211-274p24.4 |
| chr6_-_21873266 | 0.01 |
ENSDART00000151658
ENSDART00000151152 ENSDART00000151179 |
si:dkey-19e4.5
|
si:dkey-19e4.5 |
| chr3_+_40407352 | 0.01 |
ENSDART00000155112
|
tnrc18
|
trinucleotide repeat containing 18 |
| chr1_-_22756898 | 0.01 |
ENSDART00000158915
|
prom1b
|
prominin 1 b |
| chr16_-_11928774 | 0.01 |
ENSDART00000183229
|
cd4-1
|
CD4-1 molecule |
| chr12_+_27127139 | 0.01 |
ENSDART00000025966
|
hoxb6b
|
homeobox B6b |
| chr2_-_59285407 | 0.01 |
ENSDART00000181616
|
ftr34
|
finTRIM family, member 34 |
| chr25_-_31118923 | 0.01 |
ENSDART00000009126
ENSDART00000188286 |
kras
|
v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog |
| chr6_+_28140816 | 0.01 |
ENSDART00000137907
|
inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr15_-_34892664 | 0.00 |
ENSDART00000153787
ENSDART00000099721 |
rnf183
|
ring finger protein 183 |
| chr11_-_13341483 | 0.00 |
ENSDART00000164978
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr11_+_38332419 | 0.00 |
ENSDART00000005864
|
CDK18
|
si:dkey-166c18.1 |
| chr5_+_28398449 | 0.00 |
ENSDART00000165292
|
nsmfb
|
NMDA receptor synaptonuclear signaling and neuronal migration factor b |
| chr6_-_49673476 | 0.00 |
ENSDART00000112226
|
apcdd1l
|
adenomatosis polyposis coli down-regulated 1-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of tlx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.1 | 0.9 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 0.7 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 0.4 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 0.6 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.5 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 0.5 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.3 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.3 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.2 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.1 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.4 | GO:0050654 | chondroitin sulfate proteoglycan metabolic process(GO:0050654) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.1 | 0.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.2 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.1 | 0.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.4 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.8 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.0 | 0.1 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.2 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.5 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 1.0 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |