Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
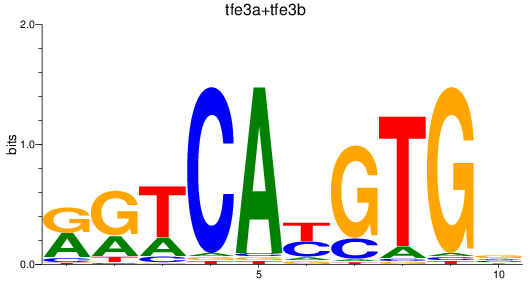
Results for tfe3a+tfe3b
Z-value: 0.49
Transcription factors associated with tfe3a+tfe3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tfe3b
|
ENSDARG00000019457 | transcription factor binding to IGHM enhancer 3b |
|
tfe3a
|
ENSDARG00000098903 | transcription factor binding to IGHM enhancer 3a |
|
tfe3b
|
ENSDARG00000111231 | transcription factor binding to IGHM enhancer 3b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tfe3a | dr11_v1_chr8_+_7778770_7778770 | -0.76 | 2.5e-04 | Click! |
| tfe3b | dr11_v1_chr11_+_25508129_25508129 | 0.44 | 6.6e-02 | Click! |
Activity profile of tfe3a+tfe3b motif
Sorted Z-values of tfe3a+tfe3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_4539899 | 2.47 |
ENSDART00000112460
|
dolk
|
dolichol kinase |
| chr13_-_3474373 | 1.86 |
ENSDART00000157437
|
prkn
|
parkin RBR E3 ubiquitin protein ligase |
| chr20_-_54014539 | 1.12 |
ENSDART00000060466
|
si:dkey-241l7.6
|
si:dkey-241l7.6 |
| chr8_-_17167819 | 1.02 |
ENSDART00000135042
ENSDART00000143920 |
mrps36
|
mitochondrial ribosomal protein S36 |
| chr15_+_1199407 | 0.83 |
ENSDART00000163827
|
mfsd1
|
major facilitator superfamily domain containing 1 |
| chr15_-_17099560 | 0.83 |
ENSDART00000101724
|
mos
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr6_+_153146 | 0.77 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr23_+_31913292 | 0.76 |
ENSDART00000136910
|
armc1l
|
armadillo repeat containing 1, like |
| chr23_+_26079467 | 0.74 |
ENSDART00000129617
|
atp6ap1b
|
ATPase H+ transporting accessory protein 1b |
| chr20_-_25631256 | 0.73 |
ENSDART00000048164
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr11_+_7183025 | 0.73 |
ENSDART00000046670
ENSDART00000154009 ENSDART00000156974 ENSDART00000125619 |
thop1
|
thimet oligopeptidase 1 |
| chr10_+_14963898 | 0.71 |
ENSDART00000187363
ENSDART00000175732 |
si:dkey-88l16.3
|
si:dkey-88l16.3 |
| chr2_-_42360329 | 0.71 |
ENSDART00000144274
ENSDART00000140863 |
si:dkey-7l6.3
|
si:dkey-7l6.3 |
| chr3_+_39568290 | 0.70 |
ENSDART00000020741
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr3_+_23029934 | 0.67 |
ENSDART00000110343
|
nags
|
N-acetylglutamate synthase |
| chr18_+_41542542 | 0.66 |
ENSDART00000087445
|
tsen34
|
TSEN34 tRNA splicing endonuclease subunit |
| chr19_+_7152966 | 0.65 |
ENSDART00000080348
|
brd2a
|
bromodomain containing 2a |
| chr21_+_4540127 | 0.65 |
ENSDART00000043431
|
nup188
|
nucleoporin 188 |
| chr17_-_25331439 | 0.64 |
ENSDART00000155422
ENSDART00000082324 |
zpcx
|
zona pellucida protein C |
| chr7_-_8324927 | 0.64 |
ENSDART00000102535
|
f13a1b
|
coagulation factor XIII, A1 polypeptide b |
| chr1_-_18811517 | 0.63 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr25_+_21716263 | 0.62 |
ENSDART00000148920
|
znf277
|
zinc finger protein 277 |
| chr1_-_47114310 | 0.61 |
ENSDART00000144899
ENSDART00000053157 |
setd4
|
SET domain containing 4 |
| chr19_-_1947403 | 0.61 |
ENSDART00000113951
ENSDART00000151293 ENSDART00000134074 |
znrf2a
|
zinc and ring finger 2a |
| chr20_+_7584211 | 0.61 |
ENSDART00000132481
ENSDART00000127975 ENSDART00000144551 |
bloc1s2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr7_-_11383933 | 0.60 |
ENSDART00000163949
|
mesd
|
mesoderm development LRP chaperone |
| chr3_+_3454610 | 0.59 |
ENSDART00000024900
|
zgc:165453
|
zgc:165453 |
| chr11_-_26590401 | 0.57 |
ENSDART00000154349
ENSDART00000123094 |
st3gal8
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 8 |
| chr11_+_42730639 | 0.56 |
ENSDART00000165297
|
zgc:194981
|
zgc:194981 |
| chr3_-_3439150 | 0.56 |
ENSDART00000021286
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr9_-_3149896 | 0.56 |
ENSDART00000020861
|
pdk1
|
pyruvate dehydrogenase kinase, isozyme 1 |
| chr12_+_34891529 | 0.55 |
ENSDART00000015643
|
tbcc
|
tubulin folding cofactor C |
| chr7_-_11384279 | 0.55 |
ENSDART00000102515
ENSDART00000172796 |
mesd
|
mesoderm development LRP chaperone |
| chr19_+_5318358 | 0.53 |
ENSDART00000082133
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr9_-_52386733 | 0.51 |
ENSDART00000171721
|
dap1b
|
death associated protein 1b |
| chr1_-_524433 | 0.50 |
ENSDART00000147610
|
si:ch73-41e3.7
|
si:ch73-41e3.7 |
| chr15_-_4580763 | 0.50 |
ENSDART00000008170
|
rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr19_-_808265 | 0.50 |
ENSDART00000082454
|
glmp
|
glycosylated lysosomal membrane protein |
| chr5_+_57210237 | 0.49 |
ENSDART00000167660
|
pja2
|
praja ring finger ubiquitin ligase 2 |
| chr19_+_167612 | 0.49 |
ENSDART00000169574
|
tatdn1
|
TatD DNase domain containing 1 |
| chr3_-_41535647 | 0.46 |
ENSDART00000153723
ENSDART00000154198 |
si:ch211-222n22.1
|
si:ch211-222n22.1 |
| chr17_-_17764801 | 0.46 |
ENSDART00000155261
|
slirp
|
SRA stem-loop interacting RNA binding protein |
| chr8_+_41229233 | 0.46 |
ENSDART00000131135
|
zgc:152830
|
zgc:152830 |
| chr3_+_34180835 | 0.46 |
ENSDART00000055252
|
timm29
|
translocase of inner mitochondrial membrane 29 |
| chr23_-_44546124 | 0.45 |
ENSDART00000126779
ENSDART00000024082 |
psmb6
|
proteasome subunit beta 6 |
| chr4_-_965267 | 0.45 |
ENSDART00000093289
|
atp23
|
ATP23 metallopeptidase and ATP synthase assembly factor homolog |
| chr9_-_23747264 | 0.44 |
ENSDART00000141461
ENSDART00000010311 |
ndufa10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10 |
| chr20_-_14665002 | 0.43 |
ENSDART00000152816
|
scrn2
|
secernin 2 |
| chr23_+_19670085 | 0.43 |
ENSDART00000031872
|
kctd6b
|
potassium channel tetramerization domain containing 6b |
| chr21_-_2958422 | 0.42 |
ENSDART00000174091
|
zgc:194215
|
zgc:194215 |
| chr18_-_1414760 | 0.41 |
ENSDART00000171881
|
PEPD
|
peptidase D |
| chr24_+_42149453 | 0.41 |
ENSDART00000128766
|
serpinb1l3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 3 |
| chr2_-_32486080 | 0.40 |
ENSDART00000110821
|
ttc19
|
tetratricopeptide repeat domain 19 |
| chr23_+_19198244 | 0.40 |
ENSDART00000047015
|
ccdc115
|
coiled-coil domain containing 115 |
| chr12_-_11649690 | 0.39 |
ENSDART00000149713
|
btbd16
|
BTB (POZ) domain containing 16 |
| chr4_-_20051141 | 0.39 |
ENSDART00000066963
|
atp6v1f
|
ATPase H+ transporting V1 subunit F |
| chr13_+_13930263 | 0.38 |
ENSDART00000079154
|
rpia
|
ribose 5-phosphate isomerase A (ribose 5-phosphate epimerase) |
| chr7_-_46019756 | 0.37 |
ENSDART00000162583
|
zgc:162297
|
zgc:162297 |
| chr3_+_43086548 | 0.37 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr1_+_52462068 | 0.37 |
ENSDART00000124682
|
glb1
|
galactosidase, beta 1 |
| chr3_+_39566999 | 0.37 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr12_-_9516981 | 0.36 |
ENSDART00000106285
|
si:ch211-207i20.3
|
si:ch211-207i20.3 |
| chr2_+_24936766 | 0.35 |
ENSDART00000025962
|
gyg1a
|
glycogenin 1a |
| chr5_-_69804433 | 0.35 |
ENSDART00000028954
|
rilpl2
|
Rab interacting lysosomal protein-like 2 |
| chr9_-_8296723 | 0.35 |
ENSDART00000139867
|
si:ch211-145c1.1
|
si:ch211-145c1.1 |
| chr13_+_28512863 | 0.34 |
ENSDART00000043117
|
fbxw4
|
F-box and WD repeat domain containing 4 |
| chr4_-_5247335 | 0.34 |
ENSDART00000050221
|
atp6v1e1b
|
ATPase H+ transporting V1 subunit E1b |
| chr15_-_1031511 | 0.34 |
ENSDART00000153569
|
si:dkey-77f5.4
|
si:dkey-77f5.4 |
| chr8_+_39767915 | 0.34 |
ENSDART00000017153
|
hps4
|
Hermansky-Pudlak syndrome 4 |
| chr15_+_37545855 | 0.33 |
ENSDART00000099456
|
psenen
|
presenilin enhancer gamma secretase subunit |
| chr20_-_13140309 | 0.33 |
ENSDART00000020703
ENSDART00000188594 |
ints7
|
integrator complex subunit 7 |
| chr10_-_309894 | 0.33 |
ENSDART00000163287
|
CABZ01049607.1
|
|
| chr23_+_31912882 | 0.33 |
ENSDART00000140505
|
armc1l
|
armadillo repeat containing 1, like |
| chr10_+_20070178 | 0.33 |
ENSDART00000027612
ENSDART00000145264 ENSDART00000172713 |
xpo7
|
exportin 7 |
| chr17_-_6535941 | 0.33 |
ENSDART00000109249
|
cenpo
|
centromere protein O |
| chr19_+_619200 | 0.32 |
ENSDART00000050125
|
nupl2
|
nucleoporin like 2 |
| chr21_+_7298687 | 0.31 |
ENSDART00000187746
|
CU929160.1
|
|
| chr13_+_28512530 | 0.31 |
ENSDART00000188510
|
fbxw4
|
F-box and WD repeat domain containing 4 |
| chr10_+_7636811 | 0.31 |
ENSDART00000160673
|
hint1
|
histidine triad nucleotide binding protein 1 |
| chr22_+_38037530 | 0.31 |
ENSDART00000012212
|
commd2
|
COMM domain containing 2 |
| chr12_+_4712215 | 0.30 |
ENSDART00000152134
|
kansl1a
|
KAT8 regulatory NSL complex subunit 1a |
| chr6_-_39198912 | 0.30 |
ENSDART00000077938
|
c1galt1b
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1b |
| chr13_-_18122333 | 0.30 |
ENSDART00000128748
|
washc2c
|
WASH complex subunit 2C |
| chr15_+_25635326 | 0.30 |
ENSDART00000135409
ENSDART00000162240 ENSDART00000052645 |
tsr1
|
TSR1, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr23_+_36340520 | 0.30 |
ENSDART00000011201
|
copz1
|
coatomer protein complex, subunit zeta 1 |
| chr5_-_41645058 | 0.30 |
ENSDART00000051092
|
riok2
|
RIO kinase 2 (yeast) |
| chr5_-_1963498 | 0.30 |
ENSDART00000073462
|
rplp0
|
ribosomal protein, large, P0 |
| chr3_+_54553931 | 0.29 |
ENSDART00000029387
|
ppan
|
peter pan homolog (Drosophila) |
| chr9_+_54686686 | 0.29 |
ENSDART00000066198
|
rab9a
|
RAB9A, member RAS oncogene family |
| chr11_-_44945636 | 0.29 |
ENSDART00000157658
|
orc2
|
origin recognition complex, subunit 2 |
| chr10_+_26118122 | 0.29 |
ENSDART00000079207
|
trim47
|
tripartite motif containing 47 |
| chr5_+_69622005 | 0.29 |
ENSDART00000167388
|
vps33a
|
vacuolar protein sorting 33A |
| chr20_-_29683754 | 0.29 |
ENSDART00000130599
ENSDART00000015928 ENSDART00000131219 |
si:ch211-195d17.2
|
si:ch211-195d17.2 |
| chr14_+_16765992 | 0.29 |
ENSDART00000140061
|
sqstm1
|
sequestosome 1 |
| chr22_+_1431743 | 0.29 |
ENSDART00000182871
|
BX322657.2
|
Danio rerio si:dkeyp-53d3.6 (si:dkeyp-53d3.6), mRNA. |
| chr19_+_7627070 | 0.29 |
ENSDART00000151078
ENSDART00000131324 |
pygo2
|
pygopus homolog 2 (Drosophila) |
| chr15_-_28596507 | 0.28 |
ENSDART00000156800
|
si:ch211-225b7.5
|
si:ch211-225b7.5 |
| chr14_+_39156 | 0.28 |
ENSDART00000082184
|
tmem107
|
transmembrane protein 107 |
| chr3_-_34180364 | 0.28 |
ENSDART00000151819
ENSDART00000003133 |
yipf2
|
Yip1 domain family, member 2 |
| chr11_-_35756468 | 0.28 |
ENSDART00000103076
|
arl8bb
|
ADP-ribosylation factor-like 8Bb |
| chr1_+_49673489 | 0.28 |
ENSDART00000135487
|
tsga10
|
testis specific, 10 |
| chr19_-_27334394 | 0.28 |
ENSDART00000052359
|
gtf2h4
|
general transcription factor IIH, polypeptide 4 |
| chr21_-_308852 | 0.28 |
ENSDART00000151613
|
lhfpl2a
|
LHFPL tetraspan subfamily member 2a |
| chr8_-_2230128 | 0.28 |
ENSDART00000140427
|
si:dkeyp-117b11.2
|
si:dkeyp-117b11.2 |
| chr23_-_22130778 | 0.28 |
ENSDART00000079212
|
phc2a
|
polyhomeotic homolog 2a (Drosophila) |
| chr23_+_384850 | 0.28 |
ENSDART00000114000
|
zgc:101663
|
zgc:101663 |
| chr8_+_23213320 | 0.27 |
ENSDART00000032996
ENSDART00000137536 |
ppdpfa
|
pancreatic progenitor cell differentiation and proliferation factor a |
| chr23_-_35066816 | 0.27 |
ENSDART00000168731
ENSDART00000163731 |
BX294434.1
|
|
| chr3_-_15859397 | 0.27 |
ENSDART00000135924
|
dhrs7b
|
dehydrogenase/reductase (SDR family) member 7B |
| chr11_-_31276064 | 0.27 |
ENSDART00000141062
ENSDART00000004780 |
man2b1
|
mannosidase, alpha, class 2B, member 1 |
| chr21_-_4950610 | 0.27 |
ENSDART00000138518
|
si:ch73-29c22.1
|
si:ch73-29c22.1 |
| chr3_+_16922226 | 0.26 |
ENSDART00000017646
|
atp6v0a1a
|
ATPase H+ transporting V0 subunit a1a |
| chr4_+_17539134 | 0.26 |
ENSDART00000013313
|
itfg2
|
integrin alpha FG-GAP repeat containing 2 |
| chr23_-_19153378 | 0.26 |
ENSDART00000019045
ENSDART00000183681 |
ebp
|
emopamil binding protein (sterol isomerase) |
| chr12_+_47044707 | 0.26 |
ENSDART00000186506
|
zranb1a
|
zinc finger, RAN-binding domain containing 1a |
| chr10_+_43037064 | 0.26 |
ENSDART00000160159
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr22_-_5661789 | 0.26 |
ENSDART00000176242
|
ccdc51
|
coiled-coil domain containing 51 |
| chr14_+_1240235 | 0.26 |
ENSDART00000127477
|
adad1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr4_+_17844013 | 0.26 |
ENSDART00000019165
|
apaf1
|
apoptotic peptidase activating factor 1 |
| chr8_+_50531709 | 0.26 |
ENSDART00000193352
|
PEBP4
|
phosphatidylethanolamine binding protein 4 |
| chr10_+_41159241 | 0.25 |
ENSDART00000141657
|
anxa4
|
annexin A4 |
| chr21_-_45891262 | 0.25 |
ENSDART00000169816
|
galnt10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
| chr18_+_14693682 | 0.25 |
ENSDART00000132249
|
uri1
|
URI1, prefoldin-like chaperone |
| chr15_+_6652396 | 0.25 |
ENSDART00000192813
ENSDART00000157678 |
nop53
|
NOP53 ribosome biogenesis factor |
| chr18_+_31280984 | 0.24 |
ENSDART00000170285
ENSDART00000150608 ENSDART00000159720 |
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr16_-_5143124 | 0.24 |
ENSDART00000131876
ENSDART00000060630 |
ttk
|
ttk protein kinase |
| chr22_+_29991834 | 0.24 |
ENSDART00000147728
|
si:dkey-286j15.3
|
si:dkey-286j15.3 |
| chr3_+_23752150 | 0.24 |
ENSDART00000146636
|
hoxb2a
|
homeobox B2a |
| chr6_+_59944488 | 0.24 |
ENSDART00000161158
|
nufip1
|
nuclear fragile X mental retardation protein interacting protein 1 |
| chr10_-_35149513 | 0.24 |
ENSDART00000063434
ENSDART00000131291 |
ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr8_+_26410197 | 0.23 |
ENSDART00000145836
ENSDART00000053447 |
ifrd2
|
interferon-related developmental regulator 2 |
| chr1_+_34496855 | 0.23 |
ENSDART00000012873
|
klf12a
|
Kruppel-like factor 12a |
| chr1_+_42345532 | 0.23 |
ENSDART00000143871
|
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr2_+_20868286 | 0.23 |
ENSDART00000062591
|
odr4
|
odr-4 GPCR localization factor homolog |
| chr11_-_44163164 | 0.23 |
ENSDART00000047126
|
clcn4
|
chloride channel, voltage-sensitive 4 |
| chr2_-_53896300 | 0.23 |
ENSDART00000161221
|
capsla
|
calcyphosine-like a |
| chr5_-_13167097 | 0.23 |
ENSDART00000149700
ENSDART00000030213 |
mapk1
|
mitogen-activated protein kinase 1 |
| chr14_+_8456870 | 0.23 |
ENSDART00000007738
|
tmco6
|
transmembrane and coiled-coil domains 6 |
| chr25_-_23583101 | 0.23 |
ENSDART00000149107
ENSDART00000103704 ENSDART00000184903 |
nap1l4a
|
nucleosome assembly protein 1-like 4a |
| chr9_+_30108641 | 0.23 |
ENSDART00000060174
|
jagn1a
|
jagunal homolog 1a |
| chr9_+_42066030 | 0.23 |
ENSDART00000185311
ENSDART00000015267 |
pcbp3
|
poly(rC) binding protein 3 |
| chr3_-_21106093 | 0.23 |
ENSDART00000156566
|
maza
|
MYC-associated zinc finger protein a (purine-binding transcription factor) |
| chr16_+_19029297 | 0.23 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr18_-_5103931 | 0.22 |
ENSDART00000188091
|
pdcd10a
|
programmed cell death 10a |
| chr25_+_16098620 | 0.22 |
ENSDART00000142564
ENSDART00000165598 |
far1
|
fatty acyl CoA reductase 1 |
| chr14_-_17575764 | 0.22 |
ENSDART00000123145
|
rnf4
|
ring finger protein 4 |
| chr5_-_22130937 | 0.22 |
ENSDART00000138606
|
las1l
|
LAS1-like, ribosome biogenesis factor |
| chr4_-_73447058 | 0.22 |
ENSDART00000172042
|
si:ch73-120g24.4
|
si:ch73-120g24.4 |
| chr13_-_18119696 | 0.22 |
ENSDART00000148125
|
washc2c
|
WASH complex subunit 2C |
| chr10_-_29768556 | 0.22 |
ENSDART00000052787
|
vps11
|
vacuolar protein sorting 11 |
| chr19_-_5103313 | 0.22 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr25_+_10547228 | 0.22 |
ENSDART00000067678
|
zgc:110339
|
zgc:110339 |
| chr7_-_41812355 | 0.22 |
ENSDART00000016105
|
vps35
|
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chr14_+_33413980 | 0.21 |
ENSDART00000052780
ENSDART00000124437 ENSDART00000173327 |
ndufa1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1 |
| chr2_-_193707 | 0.21 |
ENSDART00000187642
|
znf1014
|
zinc finger protein 1014 |
| chr7_-_41812015 | 0.21 |
ENSDART00000174058
|
vps35
|
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chr1_+_47091468 | 0.21 |
ENSDART00000036783
|
cryzl1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr25_-_14424406 | 0.21 |
ENSDART00000073609
|
prmt7
|
protein arginine methyltransferase 7 |
| chr25_+_20715950 | 0.21 |
ENSDART00000180223
|
ergic2
|
ERGIC and golgi 2 |
| chr18_-_11729 | 0.21 |
ENSDART00000159781
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr12_-_30359498 | 0.21 |
ENSDART00000152981
ENSDART00000189988 |
tdrd1
|
tudor domain containing 1 |
| chr4_+_17843717 | 0.21 |
ENSDART00000113507
|
apaf1
|
apoptotic peptidase activating factor 1 |
| chr14_+_22076596 | 0.21 |
ENSDART00000106147
ENSDART00000100278 ENSDART00000131489 |
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr16_+_4654333 | 0.21 |
ENSDART00000167665
|
LIN28A
|
si:ch1073-284b18.2 |
| chr13_-_280827 | 0.21 |
ENSDART00000144819
|
slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr13_-_280652 | 0.20 |
ENSDART00000193627
|
slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr10_-_29744921 | 0.20 |
ENSDART00000088605
|
ift46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr1_-_8566567 | 0.20 |
ENSDART00000114613
|
ptcd1
|
pentatricopeptide repeat domain 1 |
| chr14_+_52571134 | 0.20 |
ENSDART00000166708
|
rpl26
|
ribosomal protein L26 |
| chr7_-_19923249 | 0.20 |
ENSDART00000078694
|
zgc:110591
|
zgc:110591 |
| chr24_-_26854032 | 0.20 |
ENSDART00000087991
|
fndc3bb
|
fibronectin type III domain containing 3Bb |
| chr20_-_37813863 | 0.20 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr23_-_26784736 | 0.20 |
ENSDART00000024064
ENSDART00000131615 |
galnt6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) |
| chr17_+_24809221 | 0.20 |
ENSDART00000082251
ENSDART00000147871 ENSDART00000130871 |
spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr2_-_44971551 | 0.20 |
ENSDART00000018818
|
mul1a
|
mitochondrial E3 ubiquitin protein ligase 1a |
| chr20_-_40720458 | 0.20 |
ENSDART00000153151
ENSDART00000061261 ENSDART00000138569 |
cx43
|
connexin 43 |
| chr21_-_28737320 | 0.20 |
ENSDART00000098696
ENSDART00000124826 ENSDART00000125652 |
nrg2a
|
neuregulin 2a |
| chr14_+_14836468 | 0.20 |
ENSDART00000166728
|
si:dkey-102m7.3
|
si:dkey-102m7.3 |
| chr3_-_13461056 | 0.20 |
ENSDART00000137678
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr13_-_25842074 | 0.20 |
ENSDART00000015154
|
papolg
|
poly(A) polymerase gamma |
| chr17_+_44441042 | 0.20 |
ENSDART00000142123
|
ap5m1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr17_+_37215820 | 0.19 |
ENSDART00000104009
|
slc30a1b
|
solute carrier family 30 (zinc transporter), member 1b |
| chr6_-_28222592 | 0.19 |
ENSDART00000131126
|
bcl6a
|
B-cell CLL/lymphoma 6a (zinc finger protein 51) |
| chr18_-_29898351 | 0.19 |
ENSDART00000138533
ENSDART00000064076 |
cmc2
|
C-x(9)-C motif containing 2 |
| chr18_-_16924221 | 0.19 |
ENSDART00000122102
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr5_+_69950882 | 0.19 |
ENSDART00000097359
|
dnajc25
|
DnaJ (Hsp40) homolog, subfamily C , member 25 |
| chr21_+_19330774 | 0.19 |
ENSDART00000109412
|
helq
|
helicase, POLQ like |
| chr24_-_36270855 | 0.19 |
ENSDART00000154858
|
si:ch211-40k21.5
|
si:ch211-40k21.5 |
| chr18_-_898870 | 0.19 |
ENSDART00000151777
ENSDART00000062654 |
parp6a
|
poly (ADP-ribose) polymerase family, member 6a |
| chr23_+_20513104 | 0.19 |
ENSDART00000079591
|
dpm1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr19_+_3842891 | 0.19 |
ENSDART00000159043
|
lsm10
|
LSM10, U7 small nuclear RNA associated |
| chr4_-_25836684 | 0.19 |
ENSDART00000142491
|
ndufa12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr1_+_17695426 | 0.19 |
ENSDART00000103236
|
ankrd37
|
ankyrin repeat domain 37 |
| chr20_-_6476705 | 0.18 |
ENSDART00000077095
|
trappc8
|
trafficking protein particle complex 8 |
| chr25_-_6223567 | 0.18 |
ENSDART00000067512
|
psma4
|
proteasome subunit alpha 4 |
| chr24_-_7632187 | 0.18 |
ENSDART00000041714
|
atp6v0a1b
|
ATPase H+ transporting V0 subunit a1b |
| chr24_-_16917086 | 0.18 |
ENSDART00000110715
|
cmbl
|
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chr10_-_17587832 | 0.18 |
ENSDART00000113101
|
smarcad1b
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 b |
| chr16_+_35536075 | 0.18 |
ENSDART00000183618
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr2_+_33189582 | 0.18 |
ENSDART00000145588
ENSDART00000136330 ENSDART00000139295 ENSDART00000086340 |
rnf220a
|
ring finger protein 220a |
Network of associatons between targets according to the STRING database.
First level regulatory network of tfe3a+tfe3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:1905132 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.2 | 1.2 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.2 | 1.1 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.2 | 0.7 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.2 | 1.9 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.2 | 0.7 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.4 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.3 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.5 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.1 | 0.4 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.2 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 0.2 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.1 | 0.6 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.4 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 | 0.2 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.1 | 0.7 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.5 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 0.5 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.2 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.1 | 0.4 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.2 | GO:0010312 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.1 | 0.2 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 0.4 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.1 | 0.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.3 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.5 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 1.1 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.4 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.2 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.6 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.3 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 0.6 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.3 | GO:0042987 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.0 | 0.5 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.1 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.0 | 0.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 1.2 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.3 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.3 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.6 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0060343 | ventricular trabecula myocardium morphogenesis(GO:0003222) trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) skin epidermis development(GO:0098773) |
| 0.0 | 0.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.1 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.3 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.3 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.3 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.3 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.4 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 0.1 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 0.1 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.0 | 0.5 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.1 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.9 | GO:0043648 | dicarboxylic acid metabolic process(GO:0043648) |
| 0.0 | 0.1 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.2 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.0 | GO:0061178 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.4 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.1 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.0 | 0.1 | GO:1990544 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.0 | 0.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:0060416 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.5 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.1 | GO:0055016 | hypochord development(GO:0055016) |
| 0.0 | 0.1 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.0 | GO:0032677 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.5 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 0.0 | GO:0002676 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.0 | 1.6 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.6 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.2 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.2 | 0.5 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.2 | 0.7 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.2 | 0.6 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.4 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 0.7 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 0.5 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 0.3 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 0.3 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.1 | 0.2 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.1 | 0.2 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.1 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.5 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.4 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.7 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.3 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.7 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.2 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.8 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.7 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 2.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.1 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.0 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.0 | GO:0070209 | ASTRA complex(GO:0070209) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0034618 | arginine binding(GO:0034618) |
| 0.2 | 0.7 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 0.7 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.5 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 0.4 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.4 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.3 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 0.6 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.3 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 0.4 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.3 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.1 | 0.6 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.2 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.5 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 1.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.2 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 1.2 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.0 | 0.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.1 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.0 | 0.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.3 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.2 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.3 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.1 | GO:0034338 | short-chain carboxylesterase activity(GO:0034338) |
| 0.0 | 0.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.0 | 0.7 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 0.5 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.0 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.8 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 0.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.5 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.2 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.6 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.1 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |