Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
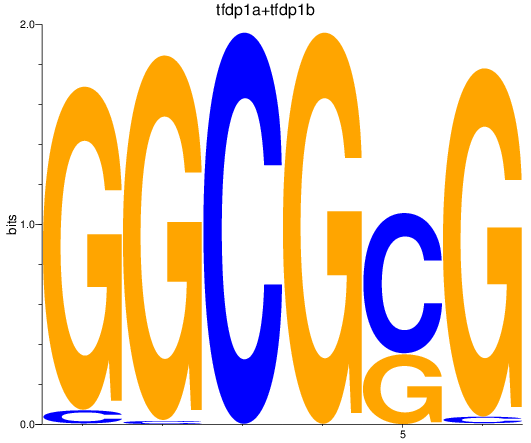
Results for tfdp1a+tfdp1b
Z-value: 1.61
Transcription factors associated with tfdp1a+tfdp1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tfdp1b
|
ENSDARG00000016304 | transcription factor Dp-1, b |
|
tfdp1a
|
ENSDARG00000019293 | transcription factor Dp-1, a |
|
tfdp1a
|
ENSDARG00000111589 | transcription factor Dp-1, a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tfdp1a | dr11_v1_chr9_+_34952203_34952269 | 0.75 | 3.1e-04 | Click! |
| tfdp1b | dr11_v1_chr1_+_227241_227241 | -0.50 | 3.5e-02 | Click! |
Activity profile of tfdp1a+tfdp1b motif
Sorted Z-values of tfdp1a+tfdp1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_1219815 | 4.20 |
ENSDART00000016800
ENSDART00000149969 |
znf367
|
zinc finger protein 367 |
| chr19_-_31007417 | 3.70 |
ENSDART00000048144
|
rbbp4
|
retinoblastoma binding protein 4 |
| chr13_-_36844945 | 3.64 |
ENSDART00000129562
ENSDART00000150899 |
nin
|
ninein (GSK3B interacting protein) |
| chr10_-_76352 | 3.49 |
ENSDART00000186560
ENSDART00000144722 |
dyrk1aa
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, a |
| chr7_+_24881680 | 3.38 |
ENSDART00000058843
|
krcp
|
kelch repeat-containing protein |
| chr4_+_9279515 | 3.29 |
ENSDART00000048707
|
srgap1b
|
SLIT-ROBO Rho GTPase activating protein 1b |
| chr3_-_36364903 | 3.29 |
ENSDART00000028883
|
gna13b
|
guanine nucleotide binding protein (G protein), alpha 13b |
| chr19_-_11846958 | 3.21 |
ENSDART00000148516
|
ctdp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr12_-_41759686 | 3.12 |
ENSDART00000172175
ENSDART00000165152 |
ppp2r2d
|
protein phosphatase 2, regulatory subunit B, delta |
| chr12_-_26538823 | 3.09 |
ENSDART00000143213
|
acsf2
|
acyl-CoA synthetase family member 2 |
| chr24_+_41989108 | 3.08 |
ENSDART00000169725
|
zbtb14
|
zinc finger and BTB domain containing 14 |
| chr21_-_18275226 | 3.06 |
ENSDART00000126672
ENSDART00000135239 |
brd3a
|
bromodomain containing 3a |
| chr7_-_69185124 | 3.06 |
ENSDART00000182217
ENSDART00000191359 |
usp10
|
ubiquitin specific peptidase 10 |
| chr8_+_387622 | 3.03 |
ENSDART00000167361
|
pym1
|
PYM homolog 1, exon junction complex associated factor |
| chr4_+_9279784 | 3.01 |
ENSDART00000014897
|
srgap1b
|
SLIT-ROBO Rho GTPase activating protein 1b |
| chr21_-_233282 | 2.93 |
ENSDART00000157684
|
bxdc2
|
brix domain containing 2 |
| chr7_-_26518086 | 2.92 |
ENSDART00000058913
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr7_-_56766100 | 2.88 |
ENSDART00000189934
|
csnk2a2a
|
casein kinase 2, alpha prime polypeptide a |
| chr5_-_28970059 | 2.86 |
ENSDART00000191119
|
fam129bb
|
family with sequence similarity 129, member Bb |
| chr23_+_36306539 | 2.85 |
ENSDART00000053267
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr7_-_69184420 | 2.82 |
ENSDART00000168311
ENSDART00000159239 ENSDART00000161319 |
usp10
|
ubiquitin specific peptidase 10 |
| chr25_-_6049339 | 2.75 |
ENSDART00000075184
|
snx1a
|
sorting nexin 1a |
| chr23_-_3758637 | 2.68 |
ENSDART00000131536
ENSDART00000139408 ENSDART00000137826 |
hmga1a
|
high mobility group AT-hook 1a |
| chr21_+_233271 | 2.64 |
ENSDART00000171440
|
dtwd2
|
DTW domain containing 2 |
| chr15_-_25435085 | 2.54 |
ENSDART00000112079
|
tlcd2
|
TLC domain containing 2 |
| chr23_-_36305874 | 2.52 |
ENSDART00000147598
ENSDART00000146986 ENSDART00000086985 ENSDART00000133259 |
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| chr12_-_31724198 | 2.50 |
ENSDART00000153056
ENSDART00000165299 ENSDART00000137464 ENSDART00000080173 |
srsf2a
|
serine/arginine-rich splicing factor 2a |
| chr23_-_36306337 | 2.50 |
ENSDART00000142760
ENSDART00000136929 ENSDART00000143340 |
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| chr20_-_29498178 | 2.48 |
ENSDART00000152986
ENSDART00000027851 ENSDART00000152954 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr23_-_29751730 | 2.48 |
ENSDART00000056865
|
ctnnbip1
|
catenin, beta interacting protein 1 |
| chr13_-_24826607 | 2.44 |
ENSDART00000087786
ENSDART00000186951 |
slka
|
STE20-like kinase a |
| chr23_-_3759345 | 2.37 |
ENSDART00000132205
ENSDART00000137707 ENSDART00000189382 |
hmga1a
|
high mobility group AT-hook 1a |
| chr6_-_33916756 | 2.32 |
ENSDART00000137447
ENSDART00000138488 |
nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr4_-_837768 | 2.26 |
ENSDART00000185280
ENSDART00000135618 |
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr16_+_33953644 | 2.26 |
ENSDART00000164447
ENSDART00000159969 |
arid1aa
|
AT rich interactive domain 1Aa (SWI-like) |
| chr11_+_45153104 | 2.25 |
ENSDART00000159204
ENSDART00000177585 |
tk1
|
thymidine kinase 1, soluble |
| chr25_+_20272145 | 2.23 |
ENSDART00000109605
|
si:dkey-219c3.2
|
si:dkey-219c3.2 |
| chr6_+_27090800 | 2.23 |
ENSDART00000121558
|
atg4b
|
autophagy related 4B, cysteine peptidase |
| chr3_+_40409100 | 2.21 |
ENSDART00000103486
|
tnrc18
|
trinucleotide repeat containing 18 |
| chr24_-_26995164 | 2.20 |
ENSDART00000142864
|
stag1b
|
stromal antigen 1b |
| chr3_-_40054615 | 2.18 |
ENSDART00000003511
ENSDART00000102540 ENSDART00000146121 |
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr17_-_23674065 | 2.15 |
ENSDART00000104735
ENSDART00000182952 |
ptena
|
phosphatase and tensin homolog A |
| chr9_+_2393764 | 2.14 |
ENSDART00000172624
|
chn1
|
chimerin 1 |
| chr4_-_77561679 | 2.14 |
ENSDART00000180809
|
AL935186.9
|
|
| chr7_+_10911396 | 2.13 |
ENSDART00000167273
ENSDART00000081323 ENSDART00000170655 |
abhd17c
|
abhydrolase domain containing 17C |
| chr3_-_1283247 | 2.12 |
ENSDART00000149814
|
tcf20
|
transcription factor 20 |
| chr14_+_14841685 | 2.12 |
ENSDART00000158291
ENSDART00000162039 |
slbp
|
stem-loop binding protein |
| chr23_+_2740741 | 2.12 |
ENSDART00000134938
|
zgc:114123
|
zgc:114123 |
| chr23_-_33558161 | 2.11 |
ENSDART00000018301
|
itga5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr9_+_55857193 | 2.11 |
ENSDART00000160980
|
sept10
|
septin 10 |
| chr1_+_31657842 | 2.10 |
ENSDART00000057880
|
poll
|
polymerase (DNA directed), lambda |
| chr8_-_410728 | 2.09 |
ENSDART00000151255
|
trim36
|
tripartite motif containing 36 |
| chr13_+_46941930 | 2.09 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr14_-_7306983 | 2.08 |
ENSDART00000158914
|
si:ch211-51f19.1
|
si:ch211-51f19.1 |
| chr4_-_2196798 | 2.07 |
ENSDART00000110178
ENSDART00000149330 |
kcnc2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr6_+_3717613 | 2.06 |
ENSDART00000184330
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr10_+_40284003 | 2.05 |
ENSDART00000062795
ENSDART00000193825 ENSDART00000113582 |
git2b
|
G protein-coupled receptor kinase interacting ArfGAP 2b |
| chr16_+_41015163 | 2.04 |
ENSDART00000058586
|
dek
|
DEK proto-oncogene |
| chr19_-_874888 | 2.02 |
ENSDART00000007206
|
eomesa
|
eomesodermin homolog a |
| chr6_-_1187565 | 2.00 |
ENSDART00000191756
|
txnrd3
|
thioredoxin reductase 3 |
| chr9_+_2762270 | 1.99 |
ENSDART00000123342
ENSDART00000001795 ENSDART00000177563 |
sp3a
|
sp3a transcription factor |
| chr22_-_10397600 | 1.98 |
ENSDART00000181964
ENSDART00000142886 |
nisch
|
nischarin |
| chr5_-_1487256 | 1.97 |
ENSDART00000149599
ENSDART00000148411 ENSDART00000092087 ENSDART00000148464 |
golga2
|
golgin A2 |
| chr1_+_31658011 | 1.97 |
ENSDART00000192203
|
poll
|
polymerase (DNA directed), lambda |
| chr19_-_42045372 | 1.96 |
ENSDART00000144275
|
trioa
|
trio Rho guanine nucleotide exchange factor a |
| chr20_+_23947004 | 1.96 |
ENSDART00000144195
|
casp8ap2
|
caspase 8 associated protein 2 |
| chr4_+_14981854 | 1.94 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr7_+_40081630 | 1.92 |
ENSDART00000173559
|
zgc:112356
|
zgc:112356 |
| chr19_-_2115040 | 1.92 |
ENSDART00000020497
|
snx13
|
sorting nexin 13 |
| chr8_-_18899427 | 1.91 |
ENSDART00000079840
|
rorca
|
RAR-related orphan receptor C a |
| chr4_+_13586455 | 1.90 |
ENSDART00000187230
|
tnpo3
|
transportin 3 |
| chr5_+_6854345 | 1.87 |
ENSDART00000066307
|
elac1
|
elaC ribonuclease Z 1 |
| chr23_+_43718115 | 1.85 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr19_-_868187 | 1.84 |
ENSDART00000186626
|
eomesa
|
eomesodermin homolog a |
| chr21_-_14692119 | 1.83 |
ENSDART00000123047
|
ehmt1b
|
euchromatic histone-lysine N-methyltransferase 1b |
| chr7_+_41887429 | 1.82 |
ENSDART00000115090
|
gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr19_-_11015238 | 1.82 |
ENSDART00000010997
|
tpm3
|
tropomyosin 3 |
| chr14_-_46198373 | 1.81 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr13_+_40815012 | 1.80 |
ENSDART00000016960
|
prkg1a
|
protein kinase, cGMP-dependent, type Ia |
| chr21_-_22357545 | 1.80 |
ENSDART00000134320
|
skp2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr25_-_17910714 | 1.78 |
ENSDART00000191586
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr8_-_25846188 | 1.76 |
ENSDART00000128829
|
efhd2
|
EF-hand domain family, member D2 |
| chr11_+_12811906 | 1.76 |
ENSDART00000123445
|
rtel1
|
regulator of telomere elongation helicase 1 |
| chr6_-_25201810 | 1.76 |
ENSDART00000168683
|
lrrc8c
|
leucine rich repeat containing 8 VRAC subunit C |
| chr17_+_26803470 | 1.75 |
ENSDART00000023470
|
pgrmc2
|
progesterone receptor membrane component 2 |
| chr19_-_47570672 | 1.75 |
ENSDART00000112155
|
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr14_-_5407118 | 1.74 |
ENSDART00000168074
|
pcgf1
|
polycomb group ring finger 1 |
| chr21_+_38033226 | 1.74 |
ENSDART00000085728
|
klf8
|
Kruppel-like factor 8 |
| chr25_+_3104959 | 1.73 |
ENSDART00000167130
|
rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr19_+_9232676 | 1.72 |
ENSDART00000136957
|
kmt2ba
|
lysine (K)-specific methyltransferase 2Ba |
| chr6_-_1187749 | 1.72 |
ENSDART00000172544
|
txnrd3
|
thioredoxin reductase 3 |
| chr6_+_41503854 | 1.71 |
ENSDART00000136538
ENSDART00000140108 ENSDART00000084861 |
cish
|
cytokine inducible SH2-containing protein |
| chr16_-_55259199 | 1.70 |
ENSDART00000161130
|
iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr6_+_41554794 | 1.70 |
ENSDART00000165424
|
srgap3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr4_+_13586689 | 1.70 |
ENSDART00000067161
ENSDART00000138201 |
tnpo3
|
transportin 3 |
| chr17_+_17804752 | 1.70 |
ENSDART00000123350
|
sptlc2a
|
serine palmitoyltransferase, long chain base subunit 2a |
| chr15_-_1622468 | 1.70 |
ENSDART00000149008
ENSDART00000034456 |
kpna4
|
karyopherin alpha 4 (importin alpha 3) |
| chr4_-_77557279 | 1.69 |
ENSDART00000180113
|
AL935186.10
|
|
| chr5_+_25760112 | 1.68 |
ENSDART00000088011
ENSDART00000182046 |
tmem2
|
transmembrane protein 2 |
| chr24_-_42072886 | 1.67 |
ENSDART00000171389
|
CABZ01095370.1
|
|
| chr4_-_77563411 | 1.65 |
ENSDART00000186841
|
AL935186.8
|
|
| chr17_-_24879003 | 1.65 |
ENSDART00000123147
|
zbtb8a
|
zinc finger and BTB domain containing 8A |
| chr12_-_37299646 | 1.64 |
ENSDART00000146142
ENSDART00000085201 |
pmp22b
|
peripheral myelin protein 22b |
| chr2_+_30739680 | 1.62 |
ENSDART00000101861
|
tcea1
|
transcription elongation factor A (SII), 1 |
| chr4_-_7869731 | 1.61 |
ENSDART00000067339
|
mcm10
|
minichromosome maintenance 10 replication initiation factor |
| chr12_+_46634736 | 1.60 |
ENSDART00000008009
|
trim16
|
tripartite motif containing 16 |
| chr14_-_246342 | 1.60 |
ENSDART00000054823
|
aurkb
|
aurora kinase B |
| chr1_-_38171648 | 1.59 |
ENSDART00000137451
ENSDART00000047159 |
hmgb2a
|
high mobility group box 2a |
| chr22_-_7025393 | 1.58 |
ENSDART00000003422
|
smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr2_+_31942390 | 1.58 |
ENSDART00000138684
ENSDART00000146758 ENSDART00000137921 |
otulinb
|
OTU deubiquitinase with linear linkage specificity b |
| chr15_+_45595385 | 1.57 |
ENSDART00000161937
ENSDART00000170214 ENSDART00000157450 |
atg16l1
|
ATG16 autophagy related 16-like 1 (S. cerevisiae) |
| chr6_+_296130 | 1.56 |
ENSDART00000073985
|
rbfox2
|
RNA binding fox-1 homolog 2 |
| chr7_-_48251234 | 1.55 |
ENSDART00000024062
ENSDART00000098904 |
cpeb1b
|
cytoplasmic polyadenylation element binding protein 1b |
| chr12_-_13729263 | 1.55 |
ENSDART00000078021
|
foxh1
|
forkhead box H1 |
| chr7_-_69429561 | 1.54 |
ENSDART00000127351
|
atxn1l
|
ataxin 1-like |
| chr12_+_35203091 | 1.54 |
ENSDART00000153022
|
ndst2b
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2b |
| chr13_-_35908275 | 1.53 |
ENSDART00000013961
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr22_-_11493236 | 1.53 |
ENSDART00000002691
|
tspan7b
|
tetraspanin 7b |
| chr16_+_41015781 | 1.53 |
ENSDART00000124543
|
dek
|
DEK proto-oncogene |
| chr6_+_20954400 | 1.52 |
ENSDART00000143248
ENSDART00000165806 |
stk11ip
|
serine/threonine kinase 11 interacting protein |
| chr3_+_51684963 | 1.52 |
ENSDART00000091180
ENSDART00000183711 ENSDART00000159493 |
baiap2a
|
BAI1-associated protein 2a |
| chr2_-_45135591 | 1.51 |
ENSDART00000014691
ENSDART00000123916 |
eif4e2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr17_-_23673864 | 1.51 |
ENSDART00000104738
ENSDART00000128958 |
ptena
|
phosphatase and tensin homolog A |
| chr22_-_155627 | 1.51 |
ENSDART00000110807
|
si:ch1073-335m2.2
|
si:ch1073-335m2.2 |
| chr9_+_21401189 | 1.50 |
ENSDART00000062669
|
cx30.3
|
connexin 30.3 |
| chr24_-_18685009 | 1.50 |
ENSDART00000158589
ENSDART00000176165 ENSDART00000167752 ENSDART00000181642 |
zgc:66014
|
zgc:66014 |
| chr19_-_3821678 | 1.50 |
ENSDART00000169639
|
si:dkey-206d17.12
|
si:dkey-206d17.12 |
| chr19_-_81851 | 1.49 |
ENSDART00000172319
|
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr11_-_27821 | 1.49 |
ENSDART00000158769
ENSDART00000172970 ENSDART00000173118 ENSDART00000168674 ENSDART00000163545 ENSDART00000173411 ENSDART00000172132 |
sp1
|
sp1 transcription factor |
| chr17_+_26828027 | 1.48 |
ENSDART00000042060
|
jade1
|
jade family PHD finger 1 |
| chr2_-_32262287 | 1.48 |
ENSDART00000056621
ENSDART00000039717 |
fam49ba
|
family with sequence similarity 49, member Ba |
| chr2_-_26642831 | 1.48 |
ENSDART00000132854
ENSDART00000087714 ENSDART00000132651 |
u2surp
|
U2 snRNP-associated SURP domain containing |
| chr20_+_2731436 | 1.48 |
ENSDART00000058779
ENSDART00000129870 ENSDART00000132186 ENSDART00000152727 |
syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr17_+_19630272 | 1.47 |
ENSDART00000104895
|
rgs7a
|
regulator of G protein signaling 7a |
| chr2_+_30740095 | 1.47 |
ENSDART00000186387
|
tcea1
|
transcription elongation factor A (SII), 1 |
| chr11_-_34577034 | 1.47 |
ENSDART00000133302
ENSDART00000184367 |
pfkfb4a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4a |
| chr7_+_24528430 | 1.46 |
ENSDART00000133022
|
si:dkeyp-75h12.2
|
si:dkeyp-75h12.2 |
| chr6_+_38626684 | 1.46 |
ENSDART00000086533
|
atp10a
|
ATPase phospholipid transporting 10A |
| chr8_+_50190742 | 1.46 |
ENSDART00000099863
|
slc25a37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr5_+_36895545 | 1.46 |
ENSDART00000135776
ENSDART00000147561 ENSDART00000133842 ENSDART00000051185 ENSDART00000141984 ENSDART00000136301 ENSDART00000142388 |
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr1_+_19538299 | 1.45 |
ENSDART00000109416
|
smc2
|
structural maintenance of chromosomes 2 |
| chr17_+_24109012 | 1.45 |
ENSDART00000156251
|
ehbp1
|
EH domain binding protein 1 |
| chr3_-_30885250 | 1.45 |
ENSDART00000109104
|
kmt5c
|
lysine methyltransferase 5C |
| chr23_+_4483083 | 1.45 |
ENSDART00000092389
|
nup210
|
nucleoporin 210 |
| chr1_-_54972170 | 1.44 |
ENSDART00000150548
ENSDART00000038330 |
khsrp
|
KH-type splicing regulatory protein |
| chr11_+_31324335 | 1.44 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr12_+_13091842 | 1.43 |
ENSDART00000185477
ENSDART00000181435 ENSDART00000124799 |
si:ch211-103b1.2
|
si:ch211-103b1.2 |
| chr9_+_711638 | 1.43 |
ENSDART00000191964
|
ybey
|
ybeY metallopeptidase |
| chr5_+_44805028 | 1.42 |
ENSDART00000141198
|
ctsla
|
cathepsin La |
| chr9_-_54840124 | 1.41 |
ENSDART00000137214
ENSDART00000085693 |
gpm6bb
|
glycoprotein M6Bb |
| chr2_+_23081247 | 1.41 |
ENSDART00000099702
ENSDART00000088867 |
mfsd12a
|
major facilitator superfamily domain containing 12a |
| chr12_-_9468618 | 1.40 |
ENSDART00000152737
ENSDART00000091519 |
pgap3
|
post-GPI attachment to proteins 3 |
| chr8_+_47683539 | 1.40 |
ENSDART00000190701
|
dpp9
|
dipeptidyl-peptidase 9 |
| chr23_-_45487304 | 1.39 |
ENSDART00000148889
|
znhit6
|
zinc finger HIT-type containing 6 |
| chr6_-_7776612 | 1.39 |
ENSDART00000190269
|
myh9a
|
myosin, heavy chain 9a, non-muscle |
| chr2_+_23701613 | 1.38 |
ENSDART00000047073
|
oxsr1a
|
oxidative stress responsive 1a |
| chr8_-_410199 | 1.38 |
ENSDART00000091177
ENSDART00000122979 ENSDART00000151331 ENSDART00000151155 |
trim36
|
tripartite motif containing 36 |
| chr20_-_3319642 | 1.37 |
ENSDART00000186743
ENSDART00000123096 |
marcksa
|
myristoylated alanine-rich protein kinase C substrate a |
| chr9_-_746317 | 1.36 |
ENSDART00000129632
ENSDART00000130720 |
usp37
|
ubiquitin specific peptidase 37 |
| chr17_-_17447899 | 1.36 |
ENSDART00000156928
ENSDART00000109034 |
nrxn3a
|
neurexin 3a |
| chr15_-_25094026 | 1.36 |
ENSDART00000129154
|
exo5
|
exonuclease 5 |
| chr2_+_27652386 | 1.35 |
ENSDART00000188261
|
tmem68
|
transmembrane protein 68 |
| chr19_-_82504 | 1.35 |
ENSDART00000027864
ENSDART00000160560 |
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr18_+_35842933 | 1.34 |
ENSDART00000151587
ENSDART00000131121 |
ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr8_+_8671229 | 1.34 |
ENSDART00000131963
|
usp11
|
ubiquitin specific peptidase 11 |
| chr23_-_43718067 | 1.33 |
ENSDART00000015777
|
abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr8_+_21225064 | 1.33 |
ENSDART00000129210
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr18_-_5248365 | 1.33 |
ENSDART00000082506
ENSDART00000082504 ENSDART00000097960 |
myef2
|
myelin expression factor 2 |
| chr2_+_23081402 | 1.32 |
ENSDART00000183073
|
mfsd12a
|
major facilitator superfamily domain containing 12a |
| chr14_-_24277805 | 1.32 |
ENSDART00000054243
|
dpf2l
|
D4, zinc and double PHD fingers family 2, like |
| chr21_-_13784859 | 1.32 |
ENSDART00000024720
|
si:ch211-282j22.3
|
si:ch211-282j22.3 |
| chr5_-_9678075 | 1.31 |
ENSDART00000097217
|
FAM109B
|
si:ch211-193c2.2 |
| chr20_+_1316803 | 1.31 |
ENSDART00000152586
ENSDART00000152165 |
nup43
|
nucleoporin 43 |
| chr7_+_13491452 | 1.31 |
ENSDART00000053535
|
arih1l
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 like |
| chr17_-_26537928 | 1.31 |
ENSDART00000155692
ENSDART00000122366 |
ccser2a
|
coiled-coil serine-rich protein 2a |
| chr5_+_13521081 | 1.31 |
ENSDART00000171975
|
si:ch211-230g14.6
|
si:ch211-230g14.6 |
| chr10_-_7821686 | 1.31 |
ENSDART00000121531
|
mat2aa
|
methionine adenosyltransferase II, alpha a |
| chr17_-_7792376 | 1.30 |
ENSDART00000064655
|
zbtb2a
|
zinc finger and BTB domain containing 2a |
| chr10_+_15255012 | 1.28 |
ENSDART00000023766
|
vldlr
|
very low density lipoprotein receptor |
| chr6_-_10037207 | 1.28 |
ENSDART00000179701
|
nudt15
|
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr18_-_20458412 | 1.27 |
ENSDART00000012241
|
kif23
|
kinesin family member 23 |
| chr3_-_2613990 | 1.27 |
ENSDART00000137102
|
si:dkey-217f16.6
|
si:dkey-217f16.6 |
| chr24_-_41267184 | 1.27 |
ENSDART00000063504
|
xylb
|
xylulokinase homolog (H. influenzae) |
| chr19_-_11031145 | 1.27 |
ENSDART00000151375
ENSDART00000027598 ENSDART00000137865 ENSDART00000188025 |
tpm3
|
tropomyosin 3 |
| chr10_+_15255198 | 1.26 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr6_+_36844127 | 1.26 |
ENSDART00000020505
|
chd1l
|
chromodomain helicase DNA binding protein 1-like |
| chr3_+_35005730 | 1.26 |
ENSDART00000029451
|
prkcbb
|
protein kinase C, beta b |
| chr2_+_47471647 | 1.25 |
ENSDART00000184199
|
acsl3b
|
acyl-CoA synthetase long chain family member 3b |
| chr6_-_6258451 | 1.24 |
ENSDART00000081966
ENSDART00000125918 |
rtn4a
|
reticulon 4a |
| chr2_-_57076687 | 1.23 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr6_+_38626926 | 1.22 |
ENSDART00000190339
|
atp10a
|
ATPase phospholipid transporting 10A |
| chr20_-_53949798 | 1.22 |
ENSDART00000153435
|
ppp2r5cb
|
protein phosphatase 2, regulatory subunit B', gamma b |
| chr2_-_32513538 | 1.22 |
ENSDART00000056640
|
abcf2a
|
ATP-binding cassette, sub-family F (GCN20), member 2a |
| chr12_-_7234915 | 1.21 |
ENSDART00000048866
|
ipmkb
|
inositol polyphosphate multikinase b |
| chr16_+_23531583 | 1.21 |
ENSDART00000146708
|
adar
|
adenosine deaminase, RNA-specific |
| chr2_-_38225388 | 1.21 |
ENSDART00000146485
ENSDART00000128043 |
acin1a
|
apoptotic chromatin condensation inducer 1a |
| chr16_+_25296389 | 1.20 |
ENSDART00000114528
|
tbc1d31
|
TBC1 domain family, member 31 |
| chr22_-_20924747 | 1.20 |
ENSDART00000185845
ENSDART00000179672 |
ell
|
elongation factor RNA polymerase II |
| chr19_+_31585341 | 1.20 |
ENSDART00000052185
|
gmnn
|
geminin, DNA replication inhibitor |
| chr5_+_36895860 | 1.20 |
ENSDART00000134493
|
srsf7a
|
serine/arginine-rich splicing factor 7a |
Network of associatons between targets according to the STRING database.
First level regulatory network of tfdp1a+tfdp1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0055109 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 1.1 | 3.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.8 | 3.3 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.8 | 4.1 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.8 | 3.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.8 | 3.0 | GO:0042779 | tRNA 3'-trailer cleavage(GO:0042779) |
| 0.7 | 2.2 | GO:0046104 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.7 | 3.6 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.7 | 2.1 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) mediolateral intercalation(GO:0060031) |
| 0.6 | 1.9 | GO:0010664 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.6 | 2.5 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.5 | 2.7 | GO:0043476 | pigment accumulation(GO:0043476) |
| 0.5 | 2.1 | GO:1904667 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.5 | 1.6 | GO:0001783 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.5 | 1.4 | GO:0034773 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.5 | 1.4 | GO:1903792 | negative regulation of anion transport(GO:1903792) |
| 0.5 | 2.7 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.4 | 1.8 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.4 | 2.6 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.4 | 1.3 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.4 | 1.7 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.4 | 2.3 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.4 | 1.2 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.4 | 1.2 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.4 | 4.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.4 | 1.5 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.4 | 1.5 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.4 | 1.5 | GO:0048322 | axial mesodermal cell differentiation(GO:0048321) axial mesodermal cell fate commitment(GO:0048322) axial mesodermal cell fate specification(GO:0048327) |
| 0.4 | 5.1 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.4 | 2.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.4 | 1.1 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.3 | 1.7 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.3 | 2.0 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.3 | 1.3 | GO:2000815 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.3 | 1.6 | GO:0039694 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.3 | 1.6 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.3 | 0.9 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.3 | 2.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.3 | 1.2 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.3 | 1.2 | GO:0010759 | regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) positive regulation of lamellipodium organization(GO:1902745) |
| 0.3 | 0.8 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.3 | 1.4 | GO:0061439 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.3 | 2.7 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.3 | 0.8 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.3 | 0.8 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.3 | 0.8 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.3 | 1.5 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.2 | 1.2 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.2 | 1.2 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.2 | 0.9 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) |
| 0.2 | 0.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 1.8 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 1.4 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.2 | 1.6 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.2 | 1.1 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.2 | 1.1 | GO:0097201 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.2 | 0.7 | GO:0071042 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.2 | 0.6 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 0.6 | GO:0051096 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.2 | 0.6 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.2 | 4.5 | GO:0030330 | DNA damage response, signal transduction by p53 class mediator(GO:0030330) |
| 0.2 | 1.4 | GO:0032205 | negative regulation of telomere maintenance(GO:0032205) |
| 0.2 | 1.4 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.2 | 0.6 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.2 | 0.6 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.2 | 1.5 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 | 1.3 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.2 | 0.7 | GO:0042148 | strand invasion(GO:0042148) |
| 0.2 | 1.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.2 | 0.5 | GO:0050428 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.2 | 1.6 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.2 | 2.2 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 0.9 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.2 | 1.0 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.2 | 1.0 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 0.2 | 1.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.2 | 2.2 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.2 | 2.1 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.2 | 1.0 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.2 | 1.0 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 | 1.3 | GO:0046070 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.2 | 1.7 | GO:0021707 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 0.8 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.2 | 0.8 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.2 | 1.7 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.2 | 1.4 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 8.1 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 0.7 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 | 4.1 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 1.5 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 1.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 3.7 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 0.8 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.8 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 1.3 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 2.9 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.5 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.1 | 2.9 | GO:2000779 | regulation of double-strand break repair(GO:2000779) |
| 0.1 | 1.7 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.9 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.1 | 2.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.4 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.1 | 0.8 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 1.2 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 1.7 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 0.6 | GO:0032979 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 2.6 | GO:0010833 | telomere maintenance via telomere lengthening(GO:0010833) |
| 0.1 | 7.4 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.1 | 0.5 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 1.1 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.1 | 2.7 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 0.4 | GO:0015990 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.1 | 0.7 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 0.9 | GO:0071333 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.1 | 1.1 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) |
| 0.1 | 0.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 2.3 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.1 | 1.2 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 1.6 | GO:0006003 | fructose metabolic process(GO:0006000) fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 2.9 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 1.1 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 3.1 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.1 | 2.0 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 3.3 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.1 | 0.6 | GO:0032366 | intracellular sterol transport(GO:0032366) |
| 0.1 | 0.9 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.1 | 0.3 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.1 | 0.3 | GO:0010898 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 1.3 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 2.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 1.5 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.9 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.8 | GO:0003315 | heart rudiment formation(GO:0003315) |
| 0.1 | 0.2 | GO:0060827 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.1 | 1.4 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 0.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 1.2 | GO:0050684 | regulation of mRNA processing(GO:0050684) regulation of mRNA metabolic process(GO:1903311) |
| 0.1 | 0.5 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.7 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.1 | 0.6 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 0.5 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.9 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 1.3 | GO:0033750 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.1 | 1.0 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 0.6 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 1.8 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.1 | 1.1 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 0.6 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.3 | GO:0048909 | dorsal spinal cord development(GO:0021516) anterior lateral line nerve development(GO:0048909) |
| 0.1 | 0.3 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 1.1 | GO:0032200 | telomere maintenance(GO:0000723) telomere organization(GO:0032200) |
| 0.1 | 1.3 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.5 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.1 | 0.2 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.1 | 0.7 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 1.0 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.7 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.1 | 1.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 0.2 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.5 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.1 | 2.8 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.1 | 1.2 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.1 | 1.0 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.7 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.1 | 0.6 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 1.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 0.9 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 1.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.4 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.1 | 2.8 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 0.9 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 1.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.3 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 0.1 | 1.0 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.7 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.9 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 2.2 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.9 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 2.1 | GO:0016571 | histone methylation(GO:0016571) |
| 0.0 | 5.1 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.4 | GO:0071684 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 1.6 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 1.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.8 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.5 | GO:0048041 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.0 | 0.2 | GO:0048521 | negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.0 | 0.9 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 1.2 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 14.2 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 2.8 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 1.1 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 | 0.8 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 1.7 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.9 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.2 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.0 | 0.9 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 2.2 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 1.2 | GO:0048538 | thymus development(GO:0048538) |
| 0.0 | 0.3 | GO:0035794 | positive regulation of mitochondrial membrane permeability(GO:0035794) mitochondrial outer membrane permeabilization(GO:0097345) positive regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902110) mitochondrial outer membrane permeabilization involved in programmed cell death(GO:1902686) |
| 0.0 | 2.3 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.1 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.0 | 0.6 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.8 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 3.4 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.8 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.6 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 1.3 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 2.2 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 0.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 1.3 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 2.1 | GO:1902284 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.0 | 0.9 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.9 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.7 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 1.7 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.2 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 0.4 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.7 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 1.3 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 1.2 | GO:0014904 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 2.6 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.5 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.0 | 1.7 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.5 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.2 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 1.5 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.0 | 0.5 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 1.1 | GO:0005977 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 1.2 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.9 | GO:0032956 | regulation of actin cytoskeleton organization(GO:0032956) |
| 0.0 | 0.2 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.4 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 1.0 | GO:0051090 | regulation of sequence-specific DNA binding transcription factor activity(GO:0051090) |
| 0.0 | 0.1 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 0.3 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.3 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 1.0 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.2 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.0 | 0.7 | GO:0042493 | response to drug(GO:0042493) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.5 | 2.0 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.4 | 1.1 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.4 | 3.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.4 | 2.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.3 | 1.0 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.3 | 1.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.3 | 3.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.3 | 1.6 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.3 | 1.6 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.3 | 3.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.3 | 2.3 | GO:0035060 | brahma complex(GO:0035060) |
| 0.3 | 2.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.3 | 1.6 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.3 | 3.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.2 | 2.7 | GO:0030904 | retromer complex(GO:0030904) |
| 0.2 | 2.5 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.2 | 1.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.2 | 0.6 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.2 | 0.8 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.2 | 2.5 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.2 | 1.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 0.7 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.2 | 2.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.2 | 1.7 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.2 | 1.1 | GO:0030892 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.2 | 6.0 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 4.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.4 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 1.1 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 1.8 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.9 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 2.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 1.1 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 1.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 2.3 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.1 | 0.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 0.4 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 1.5 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 2.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.4 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 0.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.9 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 1.6 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 1.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.3 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.1 | 1.4 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.1 | 1.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.9 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 1.5 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 0.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 1.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.2 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 1.6 | GO:1990752 | microtubule end(GO:1990752) |
| 0.1 | 2.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.6 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) outer mitochondrial membrane protein complex(GO:0098799) |
| 0.1 | 2.1 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 2.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 0.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 2.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.8 | GO:0070461 | SAGA-type complex(GO:0070461) |
| 0.0 | 1.0 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 1.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.0 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.4 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 2.0 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.8 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 2.1 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 3.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 2.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.7 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.0 | 1.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.6 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.0 | 0.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 3.9 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.9 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 2.5 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.9 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.4 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.5 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.8 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 1.0 | 3.1 | GO:0047760 | medium-chain fatty acid-CoA ligase activity(GO:0031956) butyrate-CoA ligase activity(GO:0047760) |
| 1.0 | 4.1 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 1.0 | 3.0 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.9 | 2.6 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.8 | 2.5 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.7 | 3.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.6 | 4.2 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.5 | 2.1 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.5 | 1.9 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.5 | 1.4 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.4 | 1.8 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.4 | 2.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.4 | 1.7 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.4 | 3.7 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.4 | 1.2 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.4 | 2.9 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.4 | 1.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.4 | 3.9 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.4 | 2.6 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.4 | 2.6 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.4 | 1.1 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.4 | 3.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.3 | 1.0 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.3 | 1.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.3 | 1.9 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.3 | 1.8 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.3 | 0.9 | GO:0015230 | folic acid transporter activity(GO:0008517) FAD transmembrane transporter activity(GO:0015230) |
| 0.3 | 0.8 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.3 | 2.1 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.2 | 1.7 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 1.6 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.2 | 5.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 1.9 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.2 | 1.3 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.2 | 2.1 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.2 | 2.0 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.2 | 1.5 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.2 | 0.5 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.2 | 1.5 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.2 | 0.6 | GO:0032135 | DNA insertion or deletion binding(GO:0032135) |
| 0.1 | 0.6 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.1 | 0.8 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 1.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 1.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.9 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 1.2 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.9 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 2.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.9 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.4 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.1 | 1.0 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 0.7 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 1.6 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 1.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 1.0 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 0.4 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 4.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 1.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 2.2 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 1.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 2.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 2.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.9 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 1.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 1.9 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 4.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 2.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.8 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.7 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 1.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 2.0 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.1 | 1.3 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 0.3 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.1 | 0.3 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.1 | 2.1 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 1.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 12.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 1.3 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 1.8 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 1.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 2.1 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.1 | 0.8 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 1.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.5 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.1 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.8 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 6.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 0.8 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.9 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.1 | 2.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 2.0 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 1.0 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 1.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 1.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.7 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 4.9 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 2.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.8 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.0 | 1.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.5 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 1.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.6 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 1.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.4 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.6 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 14.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 1.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 2.7 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.5 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 1.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 5.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0004776 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.7 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 2.6 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 1.3 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 8.0 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 1.8 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.6 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 1.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 1.3 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.5 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.7 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 1.4 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 5.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.7 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.4 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 9.8 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 2.1 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.0 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.0 | 0.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.3 | 10.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 2.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 5.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 1.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 4.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 3.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 0.7 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 7.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 3.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 0.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 1.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 1.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 1.8 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 0.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 2.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.3 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 2.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 3.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.7 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.8 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.1 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 10.3 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.3 | 7.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.3 | 2.8 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.2 | 4.8 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.2 | 2.9 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.2 | 3.5 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.2 | 2.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 3.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 1.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 5.4 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 0.8 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 2.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 1.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 2.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 1.8 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 0.6 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 1.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.2 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 3.4 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 2.1 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 0.9 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 0.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 2.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.0 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 2.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.1 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.6 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.2 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.1 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 1.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.1 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |