Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
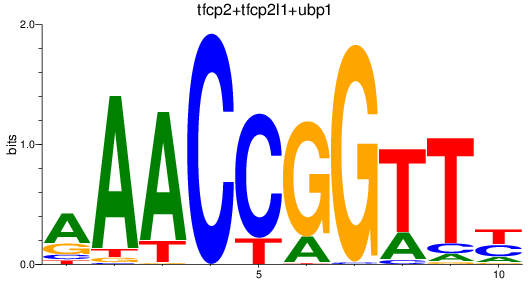
Results for tfcp2+tfcp2l1+ubp1
Z-value: 0.82
Transcription factors associated with tfcp2+tfcp2l1+ubp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ubp1
|
ENSDARG00000018000 | upstream binding protein 1 |
|
tfcp2l1
|
ENSDARG00000029497 | transcription factor CP2-like 1 |
|
tfcp2
|
ENSDARG00000060306 | transcription factor CP2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ubp1 | dr11_v1_chr19_+_42806812_42806814 | 0.92 | 9.7e-08 | Click! |
| tfcp2 | dr11_v1_chr23_-_33679579_33679579 | 0.82 | 3.0e-05 | Click! |
| tfcp2l1 | dr11_v1_chr9_+_38292947_38292947 | -0.40 | 9.6e-02 | Click! |
Activity profile of tfcp2+tfcp2l1+ubp1 motif
Sorted Z-values of tfcp2+tfcp2l1+ubp1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_51743908 | 2.20 |
ENSDART00000149039
ENSDART00000148869 |
odc1
|
ornithine decarboxylase 1 |
| chr7_+_57088920 | 1.94 |
ENSDART00000024076
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr9_-_28939181 | 1.86 |
ENSDART00000101276
ENSDART00000135334 |
epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr25_+_36292057 | 1.66 |
ENSDART00000152329
|
bmb
|
brambleberry |
| chr7_+_57089354 | 1.59 |
ENSDART00000140702
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr5_-_63065912 | 1.57 |
ENSDART00000161149
|
fam57a
|
family with sequence similarity 57 member A |
| chr25_+_36292465 | 1.57 |
ENSDART00000152649
|
bmb
|
brambleberry |
| chr15_+_12429206 | 1.55 |
ENSDART00000168997
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr16_-_6424816 | 1.53 |
ENSDART00000164864
ENSDART00000141860 |
mboat1
|
membrane bound O-acyltransferase domain containing 1 |
| chr25_+_7532627 | 1.51 |
ENSDART00000187660
|
ptdss2
|
phosphatidylserine synthase 2 |
| chr15_-_34865952 | 1.40 |
ENSDART00000186868
|
sh3bp5la
|
SH3-binding domain protein 5-like, a |
| chr15_-_34866219 | 1.36 |
ENSDART00000099723
|
sh3bp5la
|
SH3-binding domain protein 5-like, a |
| chr16_+_29509133 | 1.36 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr7_+_73670137 | 1.35 |
ENSDART00000050357
|
btr12
|
bloodthirsty-related gene family, member 12 |
| chr15_-_5901514 | 1.27 |
ENSDART00000155252
|
si:ch73-281n10.2
|
si:ch73-281n10.2 |
| chr2_-_58075414 | 1.21 |
ENSDART00000161920
|
nectin4
|
nectin cell adhesion molecule 4 |
| chr9_+_7456076 | 1.20 |
ENSDART00000125824
ENSDART00000122526 |
tmem198a
|
transmembrane protein 198a |
| chr2_-_47620806 | 1.20 |
ENSDART00000038228
|
ap1s3b
|
adaptor-related protein complex 1, sigma 3 subunit, b |
| chr1_-_53685090 | 1.19 |
ENSDART00000162751
|
xpo1a
|
exportin 1 (CRM1 homolog, yeast) a |
| chr10_+_1849874 | 1.16 |
ENSDART00000158897
ENSDART00000149956 |
apc
|
adenomatous polyposis coli |
| chr11_-_30508843 | 1.13 |
ENSDART00000101667
ENSDART00000179930 |
map4k3a
|
mitogen-activated protein kinase kinase kinase kinase 3a |
| chr5_+_26204561 | 1.13 |
ENSDART00000137178
|
marveld2b
|
MARVEL domain containing 2b |
| chr5_+_20319519 | 1.06 |
ENSDART00000004217
|
coro1ca
|
coronin, actin binding protein, 1Ca |
| chr18_+_27571448 | 1.05 |
ENSDART00000147886
|
cd82b
|
CD82 molecule b |
| chr5_-_36592307 | 1.00 |
ENSDART00000126927
ENSDART00000051196 |
taf1
|
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr24_-_28437833 | 1.00 |
ENSDART00000125412
|
fbxo45
|
F-box protein 45 |
| chr22_-_38459316 | 1.00 |
ENSDART00000149683
ENSDART00000098461 |
ptk7a
|
protein tyrosine kinase 7a |
| chr7_-_40959667 | 0.99 |
ENSDART00000084070
|
rbm33a
|
RNA binding motif protein 33a |
| chr13_-_15928934 | 0.97 |
ENSDART00000142732
|
ttl
|
tubulin tyrosine ligase |
| chr2_-_15563194 | 0.96 |
ENSDART00000049589
|
col11a1b
|
collagen, type XI, alpha 1b |
| chr10_+_36662640 | 0.96 |
ENSDART00000063359
|
ucp2
|
uncoupling protein 2 |
| chr7_+_38808027 | 0.96 |
ENSDART00000052323
|
harbi1
|
harbinger transposase derived 1 |
| chr12_-_4475890 | 0.95 |
ENSDART00000092492
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr9_-_29003245 | 0.94 |
ENSDART00000183391
ENSDART00000188836 |
ptpn4a
|
protein tyrosine phosphatase, non-receptor type 4a |
| chr13_-_24260609 | 0.93 |
ENSDART00000138747
|
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr9_-_41088279 | 0.93 |
ENSDART00000000564
|
asnsd1
|
asparagine synthetase domain containing 1 |
| chr2_+_27341559 | 0.93 |
ENSDART00000176838
|
tesk2
|
testis-specific kinase 2 |
| chr7_-_40959867 | 0.92 |
ENSDART00000174009
|
rbm33a
|
RNA binding motif protein 33a |
| chr3_-_34528306 | 0.92 |
ENSDART00000023039
|
sept9a
|
septin 9a |
| chr3_-_23512285 | 0.90 |
ENSDART00000159151
|
BX682558.1
|
|
| chr14_-_33425170 | 0.88 |
ENSDART00000124629
ENSDART00000105800 ENSDART00000001318 |
nkap
|
NFKB activating protein |
| chr8_+_29635968 | 0.88 |
ENSDART00000139029
ENSDART00000091409 |
smarcad1a
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 a |
| chr22_-_3299355 | 0.88 |
ENSDART00000190993
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr4_-_16124417 | 0.87 |
ENSDART00000128079
ENSDART00000077664 |
atp2b1a
|
ATPase plasma membrane Ca2+ transporting 1a |
| chr5_-_54712159 | 0.87 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr17_-_26537928 | 0.86 |
ENSDART00000155692
ENSDART00000122366 |
ccser2a
|
coiled-coil serine-rich protein 2a |
| chr7_+_19483277 | 0.86 |
ENSDART00000173750
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr25_+_19238175 | 0.85 |
ENSDART00000110730
ENSDART00000193619 ENSDART00000154420 |
ppip5k1b
|
diphosphoinositol pentakisphosphate kinase 1b |
| chr7_-_24838857 | 0.84 |
ENSDART00000179766
ENSDART00000180892 |
fam113
|
family with sequence similarity 113 |
| chr14_-_45558490 | 0.84 |
ENSDART00000165060
|
ints5
|
integrator complex subunit 5 |
| chr3_-_26806032 | 0.84 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr3_-_30488063 | 0.83 |
ENSDART00000055393
ENSDART00000151367 |
med25
|
mediator complex subunit 25 |
| chr12_-_28794957 | 0.83 |
ENSDART00000020667
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr7_+_13830052 | 0.83 |
ENSDART00000191360
|
abhd2a
|
abhydrolase domain containing 2a |
| chr14_-_41467497 | 0.83 |
ENSDART00000181220
|
mid1ip1l
|
MID1 interacting protein 1, like |
| chr13_+_33268657 | 0.82 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
| chr17_+_26722904 | 0.82 |
ENSDART00000114927
|
nrde2
|
NRDE-2, necessary for RNA interference, domain containing |
| chr25_+_30196039 | 0.80 |
ENSDART00000005299
|
hsd17b12a
|
hydroxysteroid (17-beta) dehydrogenase 12a |
| chr6_+_4387150 | 0.80 |
ENSDART00000181283
|
rbm26
|
RNA binding motif protein 26 |
| chr6_+_23810529 | 0.80 |
ENSDART00000166921
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr22_-_3299100 | 0.79 |
ENSDART00000160305
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr22_-_17631675 | 0.79 |
ENSDART00000132565
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr9_+_21793565 | 0.79 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr21_-_30166097 | 0.79 |
ENSDART00000130676
|
hbegfb
|
heparin-binding EGF-like growth factor b |
| chr13_-_36798204 | 0.79 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr13_+_24671481 | 0.78 |
ENSDART00000001678
|
adam8a
|
ADAM metallopeptidase domain 8a |
| chr16_+_10724406 | 0.78 |
ENSDART00000101266
|
zgc:165409
|
zgc:165409 |
| chr2_+_42871831 | 0.78 |
ENSDART00000171393
|
efr3a
|
EFR3 homolog A (S. cerevisiae) |
| chr7_-_54430505 | 0.76 |
ENSDART00000167905
|
ano1
|
anoctamin 1, calcium activated chloride channel |
| chr19_-_47570672 | 0.76 |
ENSDART00000112155
|
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr10_-_36793412 | 0.76 |
ENSDART00000185966
|
dhrs13a.2
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 2 |
| chr1_-_21321482 | 0.76 |
ENSDART00000054440
|
tmem144a
|
transmembrane protein 144a |
| chr15_-_28904371 | 0.75 |
ENSDART00000155154
|
eml2
|
echinoderm microtubule associated protein like 2 |
| chr20_-_18794789 | 0.75 |
ENSDART00000003834
|
ccm2
|
cerebral cavernous malformation 2 |
| chr14_-_31087830 | 0.74 |
ENSDART00000002250
|
hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr9_-_28939796 | 0.74 |
ENSDART00000101269
|
epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr8_-_49725430 | 0.74 |
ENSDART00000135675
|
gkap1
|
G kinase anchoring protein 1 |
| chr9_-_443451 | 0.74 |
ENSDART00000165642
|
si:dkey-11f4.14
|
si:dkey-11f4.14 |
| chr8_-_28274552 | 0.72 |
ENSDART00000131580
|
rap1aa
|
RAP1A, member of RAS oncogene family a |
| chr21_-_42831033 | 0.72 |
ENSDART00000160998
|
stk10
|
serine/threonine kinase 10 |
| chr24_-_34335265 | 0.71 |
ENSDART00000128690
|
agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr6_-_27108844 | 0.71 |
ENSDART00000073883
|
dtymk
|
deoxythymidylate kinase (thymidylate kinase) |
| chr3_-_23575007 | 0.70 |
ENSDART00000155282
ENSDART00000087726 |
igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr19_-_17304866 | 0.70 |
ENSDART00000160433
ENSDART00000190798 ENSDART00000171284 |
sf3a3
|
splicing factor 3a, subunit 3 |
| chr7_-_54217547 | 0.68 |
ENSDART00000162777
ENSDART00000188268 ENSDART00000165875 |
csnk1g1
|
casein kinase 1, gamma 1 |
| chr10_-_6587066 | 0.68 |
ENSDART00000171833
|
chd1
|
chromodomain helicase DNA binding protein 1 |
| chr20_-_25645150 | 0.68 |
ENSDART00000063137
|
si:dkeyp-117h8.4
|
si:dkeyp-117h8.4 |
| chr22_+_18349794 | 0.67 |
ENSDART00000186580
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr16_+_33992418 | 0.67 |
ENSDART00000101885
ENSDART00000130540 |
zdhhc18a
|
zinc finger, DHHC-type containing 18a |
| chr7_+_34487833 | 0.67 |
ENSDART00000173854
|
cln6a
|
CLN6, transmembrane ER protein a |
| chr23_-_18567088 | 0.66 |
ENSDART00000192371
|
sephs2
|
selenophosphate synthetase 2 |
| chr6_-_7735153 | 0.65 |
ENSDART00000151545
|
slc25a38b
|
solute carrier family 25, member 38b |
| chr12_-_13650344 | 0.65 |
ENSDART00000124364
ENSDART00000124638 ENSDART00000171929 |
stat5b
|
signal transducer and activator of transcription 5b |
| chr9_-_28255029 | 0.65 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr19_+_7173613 | 0.65 |
ENSDART00000001331
|
hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr6_-_26225814 | 0.64 |
ENSDART00000089121
|
hs2st1b
|
heparan sulfate 2-O-sulfotransferase 1b |
| chr21_-_13784859 | 0.63 |
ENSDART00000024720
|
si:ch211-282j22.3
|
si:ch211-282j22.3 |
| chr5_-_55848358 | 0.62 |
ENSDART00000130891
|
camk4
|
calcium/calmodulin-dependent protein kinase IV |
| chr2_-_7711522 | 0.62 |
ENSDART00000163175
|
dcun1d1
|
DCN1, defective in cullin neddylation 1, domain containing 1 (S. cerevisiae) |
| chr20_+_54333774 | 0.62 |
ENSDART00000144633
|
cipcb
|
CLOCK-interacting pacemaker b |
| chr22_-_34979139 | 0.62 |
ENSDART00000116455
ENSDART00000133537 |
arhgap19
|
Rho GTPase activating protein 19 |
| chr8_+_25962833 | 0.61 |
ENSDART00000086583
|
si:dkey-72l14.4
|
si:dkey-72l14.4 |
| chr20_-_28800999 | 0.61 |
ENSDART00000049462
|
rab15
|
RAB15, member RAS oncogene family |
| chr19_-_42588510 | 0.61 |
ENSDART00000102583
|
sytl1
|
synaptotagmin-like 1 |
| chr17_-_2039511 | 0.61 |
ENSDART00000160223
|
spint1a
|
serine peptidase inhibitor, Kunitz type 1 a |
| chr7_+_22792895 | 0.61 |
ENSDART00000184407
|
rbm4.3
|
RNA binding motif protein 4.3 |
| chr7_-_20021450 | 0.60 |
ENSDART00000182363
|
trip6
|
thyroid hormone receptor interactor 6 |
| chr3_+_29082267 | 0.60 |
ENSDART00000145615
|
cacna1i
|
calcium channel, voltage-dependent, T type, alpha 1I subunit |
| chr12_+_19408373 | 0.59 |
ENSDART00000114248
|
snx29
|
sorting nexin 29 |
| chr14_+_7048930 | 0.58 |
ENSDART00000109138
|
hbegfa
|
heparin-binding EGF-like growth factor a |
| chr2_+_53720028 | 0.57 |
ENSDART00000170799
|
ctnnbl1
|
catenin, beta like 1 |
| chr12_+_46634736 | 0.57 |
ENSDART00000008009
|
trim16
|
tripartite motif containing 16 |
| chr8_-_2616326 | 0.56 |
ENSDART00000027214
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr3_-_30186296 | 0.56 |
ENSDART00000134395
ENSDART00000077057 ENSDART00000017422 |
tbc1d17
|
TBC1 domain family, member 17 |
| chr16_+_46410520 | 0.55 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr1_+_46404968 | 0.55 |
ENSDART00000042064
|
tubgcp3
|
tubulin, gamma complex associated protein 3 |
| chr5_-_30418636 | 0.54 |
ENSDART00000098285
|
atf5a
|
activating transcription factor 5a |
| chr17_-_33415740 | 0.54 |
ENSDART00000135218
|
ccdc28a
|
coiled-coil domain containing 28A |
| chr22_-_14161309 | 0.54 |
ENSDART00000133365
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr19_-_2861444 | 0.53 |
ENSDART00000169053
|
clec3bb
|
C-type lectin domain family 3, member Bb |
| chr17_+_25187226 | 0.53 |
ENSDART00000148431
|
cln8
|
CLN8, transmembrane ER and ERGIC protein |
| chr5_-_67115872 | 0.53 |
ENSDART00000065262
|
rps6ka4
|
ribosomal protein S6 kinase, polypeptide 4 |
| chr9_+_21260314 | 0.52 |
ENSDART00000145943
|
ska3
|
spindle and kinetochore associated complex subunit 3 |
| chr7_+_30988570 | 0.52 |
ENSDART00000180613
ENSDART00000185625 |
tjp1a
|
tight junction protein 1a |
| chr21_+_34088377 | 0.51 |
ENSDART00000170070
|
mtmr1b
|
myotubularin related protein 1b |
| chr21_+_25801345 | 0.51 |
ENSDART00000035062
|
nf2b
|
neurofibromin 2b (merlin) |
| chr12_-_850457 | 0.51 |
ENSDART00000022688
|
tob1b
|
transducer of ERBB2, 1b |
| chr25_+_17244532 | 0.51 |
ENSDART00000050379
ENSDART00000171322 |
kdm7ab
|
lysine (K)-specific demethylase 7Ab |
| chr14_+_21755469 | 0.50 |
ENSDART00000186326
|
kdm2ab
|
lysine (K)-specific demethylase 2Ab |
| chr4_-_73488406 | 0.50 |
ENSDART00000115002
|
si:ch73-266f23.1
|
si:ch73-266f23.1 |
| chr8_-_22326744 | 0.49 |
ENSDART00000137645
|
cep104
|
centrosomal protein 104 |
| chr10_-_1523253 | 0.48 |
ENSDART00000179510
ENSDART00000176548 ENSDART00000180368 ENSDART00000185270 |
WDR70
|
WD repeat domain 70 |
| chr17_-_33416020 | 0.47 |
ENSDART00000140149
|
ccdc28a
|
coiled-coil domain containing 28A |
| chr17_+_50074372 | 0.47 |
ENSDART00000113644
|
vps39
|
vacuolar protein sorting 39 homolog (S. cerevisiae) |
| chr12_-_24812403 | 0.47 |
ENSDART00000185517
|
foxn2b
|
forkhead box N2b |
| chr21_-_40398619 | 0.47 |
ENSDART00000136068
ENSDART00000007981 ENSDART00000130950 |
nup98
|
nucleoporin 98 |
| chr23_-_17429775 | 0.46 |
ENSDART00000043076
|
ppdpfb
|
pancreatic progenitor cell differentiation and proliferation factor b |
| chr11_+_3246059 | 0.46 |
ENSDART00000161529
|
timeless
|
timeless circadian clock |
| chr12_-_21681509 | 0.46 |
ENSDART00000112726
|
eme1
|
essential meiotic structure-specific endonuclease 1 |
| chr14_-_31862657 | 0.46 |
ENSDART00000172870
ENSDART00000007927 ENSDART00000134748 ENSDART00000128730 |
rbmx
|
RNA binding motif protein, X-linked |
| chr24_+_37825634 | 0.45 |
ENSDART00000129889
|
ift140
|
intraflagellar transport 140 homolog (Chlamydomonas) |
| chr23_-_3511630 | 0.45 |
ENSDART00000019667
|
rnf114
|
ring finger protein 114 |
| chr25_+_10416583 | 0.45 |
ENSDART00000073907
|
ehf
|
ets homologous factor |
| chr14_+_45350008 | 0.44 |
ENSDART00000025749
|
ttc9c
|
tetratricopeptide repeat domain 9C |
| chr15_-_4596623 | 0.44 |
ENSDART00000132227
|
eif4h
|
eukaryotic translation initiation factor 4h |
| chr5_+_58665648 | 0.43 |
ENSDART00000167481
|
zgc:194948
|
zgc:194948 |
| chr4_-_27129697 | 0.43 |
ENSDART00000131240
|
zbed4
|
zinc finger, BED-type containing 4 |
| chr3_+_41731527 | 0.43 |
ENSDART00000049007
ENSDART00000187866 |
chst12a
|
carbohydrate (chondroitin 4) sulfotransferase 12a |
| chr7_+_54259657 | 0.43 |
ENSDART00000170174
|
pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr5_+_37966505 | 0.43 |
ENSDART00000127648
|
pafah1b2
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 |
| chr13_+_18471546 | 0.43 |
ENSDART00000090712
ENSDART00000127962 |
stox1
|
storkhead box 1 |
| chr17_-_21049260 | 0.43 |
ENSDART00000155300
ENSDART00000185758 |
bicc1a
|
BicC family RNA binding protein 1a |
| chr20_+_25645459 | 0.42 |
ENSDART00000143555
ENSDART00000036350 |
aasdh
|
aminoadipate-semialdehyde dehydrogenase |
| chr14_+_20941534 | 0.42 |
ENSDART00000185616
|
zgc:66433
|
zgc:66433 |
| chr25_+_35891342 | 0.41 |
ENSDART00000147093
|
lsm14aa
|
LSM14A mRNA processing body assembly factor a |
| chr23_-_270847 | 0.40 |
ENSDART00000191867
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr8_-_11067079 | 0.40 |
ENSDART00000181986
|
dennd2c
|
DENN/MADD domain containing 2C |
| chr10_-_15048781 | 0.40 |
ENSDART00000038401
ENSDART00000155674 |
si:ch211-95j8.2
|
si:ch211-95j8.2 |
| chr6_-_21726758 | 0.40 |
ENSDART00000083085
|
mtmr14
|
myotubularin related protein 14 |
| chr7_-_69352424 | 0.39 |
ENSDART00000170714
|
ap1g1
|
adaptor-related protein complex 1, gamma 1 subunit |
| chr2_-_9527129 | 0.39 |
ENSDART00000157422
ENSDART00000004398 |
cope
|
coatomer protein complex, subunit epsilon |
| chr6_+_149405 | 0.38 |
ENSDART00000161154
|
fdx1l
|
ferredoxin 1-like |
| chr16_-_36196700 | 0.38 |
ENSDART00000172324
|
capn7
|
calpain 7 |
| chr21_+_4256291 | 0.38 |
ENSDART00000148138
|
lrrc8aa
|
leucine rich repeat containing 8 VRAC subunit Aa |
| chr5_+_31779911 | 0.38 |
ENSDART00000098163
|
slc25a25b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25b |
| chr20_+_23083800 | 0.38 |
ENSDART00000132093
|
usp46
|
ubiquitin specific peptidase 46 |
| chr8_-_20914829 | 0.37 |
ENSDART00000025356
|
haus5
|
HAUS augmin-like complex, subunit 5 |
| chr15_-_20412286 | 0.36 |
ENSDART00000008589
|
chp2
|
calcineurin-like EF-hand protein 2 |
| chr7_+_67748939 | 0.34 |
ENSDART00000162978
|
dhx38
|
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr20_-_36416922 | 0.34 |
ENSDART00000019145
|
lbr
|
lamin B receptor |
| chr20_-_43917647 | 0.33 |
ENSDART00000026213
|
mark3b
|
MAP/microtubule affinity-regulating kinase 3b |
| chr6_+_41452979 | 0.33 |
ENSDART00000007353
|
wdr82
|
WD repeat domain 82 |
| chr3_+_24511959 | 0.33 |
ENSDART00000133898
|
dnal4a
|
dynein, axonemal, light chain 4a |
| chr3_+_16724614 | 0.33 |
ENSDART00000182135
|
gys1
|
glycogen synthase 1 (muscle) |
| chr21_-_32467799 | 0.33 |
ENSDART00000007675
ENSDART00000133099 |
zgc:123105
|
zgc:123105 |
| chr21_-_32467099 | 0.33 |
ENSDART00000186354
|
zgc:123105
|
zgc:123105 |
| chr1_-_55183088 | 0.33 |
ENSDART00000100619
|
LUC7L2
|
zgc:158803 |
| chr15_-_601971 | 0.32 |
ENSDART00000188882
|
alox5b.3
|
arachidonate 5-lipoxygenase b, tandem duplicate 3 |
| chr13_+_35528607 | 0.32 |
ENSDART00000075414
ENSDART00000112947 |
wdr27
|
WD repeat domain 27 |
| chr20_+_23408970 | 0.32 |
ENSDART00000144261
|
fryl
|
furry homolog, like |
| chr21_-_40557281 | 0.32 |
ENSDART00000172327
|
taok1b
|
TAO kinase 1b |
| chr1_-_11104805 | 0.32 |
ENSDART00000147648
|
knl1
|
kinetochore scaffold 1 |
| chr21_-_30031396 | 0.31 |
ENSDART00000157167
|
pwwp2a
|
PWWP domain containing 2A |
| chr14_+_22457230 | 0.30 |
ENSDART00000019296
|
gdf9
|
growth differentiation factor 9 |
| chr17_-_7351488 | 0.30 |
ENSDART00000098731
|
stxbp5b
|
syntaxin binding protein 5b (tomosyn) |
| chr5_+_30741730 | 0.30 |
ENSDART00000098246
ENSDART00000186992 ENSDART00000182533 |
ftr83
|
finTRIM family, member 83 |
| chr5_+_26805608 | 0.30 |
ENSDART00000111019
|
rabl6b
|
RAB, member RAS oncogene family-like 6b |
| chr18_-_40773413 | 0.29 |
ENSDART00000133797
|
vaspb
|
vasodilator stimulated phosphoprotein b |
| chr12_-_10476448 | 0.29 |
ENSDART00000106172
|
rac1a
|
Rac family small GTPase 1a |
| chr20_-_28884800 | 0.29 |
ENSDART00000134564
ENSDART00000132127 ENSDART00000135506 ENSDART00000075515 |
srsf5b
|
serine/arginine-rich splicing factor 5b |
| chr16_+_5597600 | 0.29 |
ENSDART00000017307
|
zgc:91890
|
zgc:91890 |
| chr13_-_41482064 | 0.29 |
ENSDART00000188322
ENSDART00000164732 |
pcdh15a
|
protocadherin-related 15a |
| chr8_+_7778770 | 0.29 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr10_-_24317274 | 0.28 |
ENSDART00000187956
|
inpp5kb
|
inositol polyphosphate-5-phosphatase Kb |
| chr3_-_27647845 | 0.28 |
ENSDART00000151625
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr18_+_30157198 | 0.27 |
ENSDART00000172579
|
gse1
|
Gse1 coiled-coil protein |
| chr6_+_40931018 | 0.27 |
ENSDART00000188975
|
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr6_-_24103666 | 0.27 |
ENSDART00000164915
|
scinla
|
scinderin like a |
| chr16_+_29586468 | 0.27 |
ENSDART00000148926
|
mcl1b
|
MCL1, BCL2 family apoptosis regulator b |
| chr22_+_18319230 | 0.27 |
ENSDART00000184747
ENSDART00000184649 |
gatad2ab
|
GATA zinc finger domain containing 2Ab |
Network of associatons between targets according to the STRING database.
First level regulatory network of tfcp2+tfcp2l1+ubp1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.2 | GO:0007344 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.3 | 2.6 | GO:0021550 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.3 | 1.2 | GO:1905067 | endodermal digestive tract morphogenesis(GO:0061031) Wnt signaling pathway involved in somitogenesis(GO:0090244) negative regulation of canonical Wnt signaling pathway involved in heart development(GO:1905067) |
| 0.3 | 0.8 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 2.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.2 | 0.7 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.2 | 0.9 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.2 | 2.8 | GO:0061099 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.2 | 0.7 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.2 | 0.8 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.2 | 0.8 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.2 | 0.9 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.2 | 0.8 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.2 | 0.6 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.2 | 1.4 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 | 1.4 | GO:0045741 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.2 | 1.5 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.7 | GO:1902804 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.4 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 1.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.6 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 1.0 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.1 | 1.0 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 0.8 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 0.8 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.8 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.3 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 1.0 | GO:0060386 | synapse assembly involved in innervation(GO:0060386) |
| 0.1 | 0.6 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.1 | 0.3 | GO:0030237 | female sex determination(GO:0030237) |
| 0.1 | 0.9 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 0.5 | GO:0043111 | replication fork arrest(GO:0043111) |
| 0.1 | 0.5 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 0.5 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 0.8 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 1.0 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 0.6 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.1 | 0.9 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 0.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.4 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 0.6 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.7 | GO:0014028 | notochord formation(GO:0014028) |
| 0.1 | 0.5 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.3 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.3 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.1 | 0.3 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.1 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.3 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.5 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.4 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.5 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.6 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.2 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.0 | 0.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.8 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 1.1 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.7 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 0.8 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.3 | GO:0070874 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.0 | 0.1 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.5 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.8 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.7 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.8 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.3 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 1.2 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 2.0 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 1.9 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.3 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.1 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.7 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.4 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.7 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.3 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.5 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.8 | GO:0071166 | ribonucleoprotein complex localization(GO:0071166) |
| 0.0 | 0.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.1 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.0 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.0 | GO:0060958 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.0 | 0.8 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.0 | GO:0071830 | plasma lipoprotein particle assembly(GO:0034377) high-density lipoprotein particle assembly(GO:0034380) chylomicron remnant clearance(GO:0034382) protein-lipid complex assembly(GO:0065005) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.0 | 0.3 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.3 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.4 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) synaptic vesicle budding(GO:0070142) |
| 0.0 | 0.5 | GO:0030282 | bone mineralization(GO:0030282) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.3 | 0.9 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.3 | 0.8 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.2 | 0.8 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 0.5 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.2 | 3.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 1.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.8 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.4 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.5 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 0.5 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.5 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 0.7 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 1.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.0 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 4.2 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 3.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.9 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 0.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.2 | GO:0030117 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 0.7 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.9 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.2 | 1.0 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.2 | 0.7 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.2 | 0.9 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.2 | 0.6 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.2 | 0.9 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.2 | 1.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.8 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.1 | 0.7 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 0.8 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 1.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.8 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.8 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.8 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 2.8 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.1 | 0.8 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 0.3 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.1 | 0.4 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.1 | 0.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 1.0 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 0.5 | GO:0097001 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.1 | 1.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.2 | GO:0004475 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.0 | 0.3 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.3 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.3 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 0.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.3 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.2 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.7 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.8 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.6 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 1.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.4 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 1.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.9 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.5 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 0.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 2.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.0 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.5 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.0 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.5 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 3.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.0 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.7 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 2.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.6 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 1.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 0.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.6 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 1.6 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 1.0 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |