Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
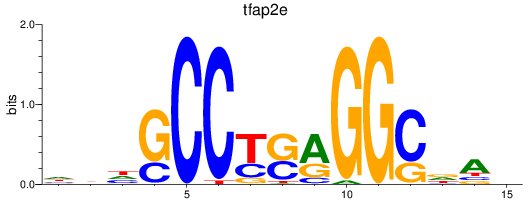
Results for tfap2e
Z-value: 0.69
Transcription factors associated with tfap2e
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tfap2e
|
ENSDARG00000008861 | transcription factor AP-2 epsilon |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tfap2e | dr11_v1_chr19_-_47456787_47456787 | -0.82 | 3.7e-05 | Click! |
Activity profile of tfap2e motif
Sorted Z-values of tfap2e motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_54607166 | 2.17 |
ENSDART00000021977
|
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr14_-_34605607 | 1.87 |
ENSDART00000191608
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr24_-_21903588 | 1.62 |
ENSDART00000180991
|
spata13
|
spermatogenesis associated 13 |
| chr24_-_21903360 | 1.60 |
ENSDART00000091252
|
spata13
|
spermatogenesis associated 13 |
| chr14_-_34605804 | 1.49 |
ENSDART00000144547
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr2_-_15041846 | 1.44 |
ENSDART00000139050
|
si:dkey-10f21.4
|
si:dkey-10f21.4 |
| chr19_+_2835240 | 1.38 |
ENSDART00000190838
|
CDCP1
|
CUB domain containing protein 1 |
| chr20_+_6533260 | 1.34 |
ENSDART00000135005
ENSDART00000166356 |
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr19_+_32979331 | 1.12 |
ENSDART00000078066
|
spire1a
|
spire-type actin nucleation factor 1a |
| chr5_+_63302660 | 1.11 |
ENSDART00000142131
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr7_+_32693890 | 1.10 |
ENSDART00000121972
|
slc39a13
|
solute carrier family 39 (zinc transporter), member 13 |
| chr23_+_27778670 | 1.07 |
ENSDART00000053863
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr2_+_1988036 | 1.02 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr8_+_48942470 | 1.00 |
ENSDART00000005464
ENSDART00000132035 |
rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr18_+_34181655 | 1.00 |
ENSDART00000130831
ENSDART00000109535 |
gmps
|
guanine monophosphate synthase |
| chr19_-_31707892 | 0.99 |
ENSDART00000088427
|
ripor2
|
RHO family interacting cell polarization regulator 2 |
| chr23_-_18668836 | 0.97 |
ENSDART00000138792
ENSDART00000051182 |
arhgap4b
|
Rho GTPase activating protein 4b |
| chr3_-_55328548 | 0.93 |
ENSDART00000082944
|
dock6
|
dedicator of cytokinesis 6 |
| chr5_+_25736979 | 0.92 |
ENSDART00000175959
|
abhd17b
|
abhydrolase domain containing 17B |
| chr13_+_16342926 | 0.87 |
ENSDART00000105496
ENSDART00000124710 |
dlg5a
|
discs, large homolog 5a (Drosophila) |
| chr7_+_39011852 | 0.85 |
ENSDART00000093009
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr1_-_25486471 | 0.82 |
ENSDART00000134200
ENSDART00000141892 ENSDART00000102501 |
arfip1
|
ADP-ribosylation factor interacting protein 1 (arfaptin 1) |
| chr18_+_50650512 | 0.81 |
ENSDART00000161022
|
si:dkey-151j17.4
|
si:dkey-151j17.4 |
| chr6_+_153146 | 0.81 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr11_+_31323746 | 0.78 |
ENSDART00000180220
ENSDART00000189937 |
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr11_+_14333441 | 0.78 |
ENSDART00000171969
|
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr8_+_48943009 | 0.75 |
ENSDART00000180763
|
rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr1_+_16548733 | 0.74 |
ENSDART00000048855
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr2_+_36616830 | 0.72 |
ENSDART00000169547
ENSDART00000098417 |
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr14_-_31465905 | 0.69 |
ENSDART00000173108
|
gpc3
|
glypican 3 |
| chr11_+_31324335 | 0.68 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr2_+_30547018 | 0.67 |
ENSDART00000193747
|
ankrd33bb
|
ankyrin repeat domain 33Bb |
| chr14_+_29609245 | 0.60 |
ENSDART00000043058
|
TENM2
|
si:dkey-34l15.2 |
| chr25_-_19574146 | 0.59 |
ENSDART00000156811
|
si:ch211-59o9.10
|
si:ch211-59o9.10 |
| chr3_+_3454610 | 0.58 |
ENSDART00000024900
|
zgc:165453
|
zgc:165453 |
| chr8_-_11004726 | 0.55 |
ENSDART00000192594
ENSDART00000020116 |
trim33
|
tripartite motif containing 33 |
| chr3_+_25990052 | 0.54 |
ENSDART00000007398
|
gcat
|
glycine C-acetyltransferase |
| chr9_-_2892045 | 0.52 |
ENSDART00000137201
|
cdca7a
|
cell division cycle associated 7a |
| chr24_+_31374324 | 0.50 |
ENSDART00000172335
ENSDART00000163162 |
cpne3
|
copine III |
| chr5_+_27267186 | 0.50 |
ENSDART00000182238
ENSDART00000087857 |
unc5db
|
unc-5 netrin receptor Db |
| chr9_-_2892250 | 0.48 |
ENSDART00000140695
|
cdca7a
|
cell division cycle associated 7a |
| chr11_+_30253968 | 0.47 |
ENSDART00000157272
ENSDART00000003475 |
ppef1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr7_+_39011355 | 0.47 |
ENSDART00000173855
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr19_-_5369486 | 0.47 |
ENSDART00000105004
|
krt17
|
keratin 17 |
| chr10_-_5135788 | 0.41 |
ENSDART00000108587
ENSDART00000138537 |
sec31a
|
SEC31 homolog A, COPII coat complex component |
| chr6_-_25369295 | 0.41 |
ENSDART00000191475
|
PKN2 (1 of many)
|
zgc:153916 |
| chr23_-_35347714 | 0.41 |
ENSDART00000161770
ENSDART00000165615 |
cpne9
|
copine family member IX |
| chr11_-_28050559 | 0.40 |
ENSDART00000136859
|
ece1
|
endothelin converting enzyme 1 |
| chr15_+_44929497 | 0.38 |
ENSDART00000111373
|
pdgfd
|
platelet derived growth factor d |
| chr12_+_19036380 | 0.37 |
ENSDART00000153086
ENSDART00000181060 |
kctd17
|
potassium channel tetramerization domain containing 17 |
| chr25_+_11281970 | 0.37 |
ENSDART00000180094
|
AKAP13
|
si:dkey-187e18.1 |
| chr14_+_26437891 | 0.34 |
ENSDART00000175459
|
gpr137
|
G protein-coupled receptor 137 |
| chr14_-_26704829 | 0.34 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr19_+_42660158 | 0.33 |
ENSDART00000018328
|
fbxl2
|
F-box and leucine-rich repeat protein 2 |
| chr7_-_15252540 | 0.33 |
ENSDART00000173072
ENSDART00000125258 |
si:dkey-172h23.2
|
si:dkey-172h23.2 |
| chr5_-_14509137 | 0.32 |
ENSDART00000180742
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr6_-_39313027 | 0.32 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr17_+_34186632 | 0.32 |
ENSDART00000014306
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr10_-_2526526 | 0.32 |
ENSDART00000191726
ENSDART00000192767 |
CU856539.1
|
|
| chr6_+_28203 | 0.31 |
ENSDART00000191561
|
CZQB01141835.1
|
|
| chr2_+_11206317 | 0.30 |
ENSDART00000144982
|
lhx8a
|
LIM homeobox 8a |
| chr22_+_15633013 | 0.24 |
ENSDART00000188095
ENSDART00000048763 |
CABZ01090041.1
|
|
| chr19_-_30510930 | 0.23 |
ENSDART00000088760
ENSDART00000181043 |
bag6l
|
BCL2 associated athanogene 6, like |
| chr4_+_77966055 | 0.23 |
ENSDART00000174203
ENSDART00000130100 ENSDART00000080665 ENSDART00000174317 ENSDART00000190123 |
zgc:113921
|
zgc:113921 |
| chr3_-_49163683 | 0.19 |
ENSDART00000166146
|
dnajb1a
|
DnaJ (Hsp40) homolog, subfamily B, member 1a |
| chr2_+_11205795 | 0.19 |
ENSDART00000019078
|
lhx8a
|
LIM homeobox 8a |
| chr23_-_4930229 | 0.19 |
ENSDART00000189456
|
ngfa
|
nerve growth factor a (beta polypeptide) |
| chr20_-_2725930 | 0.16 |
ENSDART00000081643
|
akirin2
|
akirin 2 |
| chr16_-_17162485 | 0.14 |
ENSDART00000123011
|
iffo1b
|
intermediate filament family orphan 1b |
| chr23_+_37323962 | 0.13 |
ENSDART00000102881
|
fam43b
|
family with sequence similarity 43, member B |
| chr19_-_10395683 | 0.11 |
ENSDART00000109488
|
zgc:194578
|
zgc:194578 |
| chr11_+_13067028 | 0.09 |
ENSDART00000186126
|
zfyve9b
|
zinc finger, FYVE domain containing 9b |
| chr5_-_26936861 | 0.08 |
ENSDART00000191032
|
htra4
|
HtrA serine peptidase 4 |
| chr21_+_39941875 | 0.08 |
ENSDART00000190414
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr21_+_39941559 | 0.06 |
ENSDART00000189718
ENSDART00000160875 ENSDART00000135235 |
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr16_+_36906693 | 0.06 |
ENSDART00000160645
|
si:ch73-215d9.1
|
si:ch73-215d9.1 |
| chr25_+_5249513 | 0.05 |
ENSDART00000126814
|
CABZ01039863.1
|
|
| chr18_+_34478959 | 0.05 |
ENSDART00000059394
|
kcnab1a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 a |
| chr7_-_2116512 | 0.02 |
ENSDART00000098148
|
si:cabz01007802.1
|
si:cabz01007802.1 |
| chr18_-_47103131 | 0.02 |
ENSDART00000188553
|
CABZ01039304.1
|
|
| chr22_+_9418502 | 0.01 |
ENSDART00000189702
|
si:ch211-11p18.6
|
si:ch211-11p18.6 |
| chr1_-_53476758 | 0.01 |
ENSDART00000074221
|
b3gnt2a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2a |
| chr20_-_15089738 | 0.01 |
ENSDART00000164552
|
si:dkey-239i20.2
|
si:dkey-239i20.2 |
| chr5_+_38673168 | 0.01 |
ENSDART00000135267
|
si:dkey-58f10.12
|
si:dkey-58f10.12 |
| chr4_-_1914228 | 0.01 |
ENSDART00000087835
|
ano6
|
anoctamin 6 |
| chr3_-_8160457 | 0.01 |
ENSDART00000183379
|
si:ch211-51i16.1
|
si:ch211-51i16.1 |
| chr19_+_1184878 | 0.01 |
ENSDART00000163539
|
scrt1a
|
scratch family zinc finger 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of tfap2e
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.3 | 1.0 | GO:2000402 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.3 | 1.8 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 1.1 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.1 | 0.4 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 1.1 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 1.0 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.1 | 0.7 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.1 | 1.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.3 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.1 | 0.9 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.5 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.1 | 0.3 | GO:0048909 | dorsal spinal cord development(GO:0021516) anterior lateral line nerve development(GO:0048909) |
| 0.1 | 0.5 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.1 | 1.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 1.1 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.4 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.5 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.8 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 1.0 | GO:0048538 | thymus development(GO:0048538) |
| 0.0 | 0.3 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.3 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.5 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 1.4 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.9 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.8 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 1.0 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.0 | GO:1902260 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.2 | 3.4 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 1.1 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.0 | GO:0036064 | ciliary basal body(GO:0036064) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.2 | 2.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.5 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.1 | 0.8 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 1.0 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 3.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 1.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.5 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.9 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.5 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.5 | GO:0030145 | manganese ion binding(GO:0030145) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 2.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.4 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |