Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
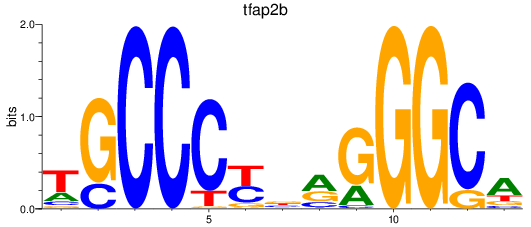
Results for tfap2b
Z-value: 0.23
Transcription factors associated with tfap2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tfap2b
|
ENSDARG00000012667 | transcription factor AP-2 beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tfap2b | dr11_v1_chr20_-_47704973_47704973 | -0.49 | 3.8e-02 | Click! |
Activity profile of tfap2b motif
Sorted Z-values of tfap2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_77551860 | 0.64 |
ENSDART00000188176
|
AL935186.6
|
|
| chr4_-_77563411 | 0.63 |
ENSDART00000186841
|
AL935186.8
|
|
| chr7_+_34794829 | 0.40 |
ENSDART00000009698
ENSDART00000075089 ENSDART00000173456 |
esrp2
|
epithelial splicing regulatory protein 2 |
| chr16_+_26612401 | 0.34 |
ENSDART00000145571
|
epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr1_-_23293261 | 0.33 |
ENSDART00000122648
|
ugdh
|
UDP-glucose 6-dehydrogenase |
| chr1_-_55248496 | 0.31 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr22_+_10781894 | 0.30 |
ENSDART00000081183
|
enc3
|
ectodermal-neural cortex 3 |
| chr23_+_25868637 | 0.29 |
ENSDART00000103934
|
ttpal
|
tocopherol (alpha) transfer protein-like |
| chr8_+_23861461 | 0.28 |
ENSDART00000037109
|
srpk1a
|
SRSF protein kinase 1a |
| chr16_+_20496691 | 0.27 |
ENSDART00000182737
ENSDART00000078984 |
cpvl
|
carboxypeptidase, vitellogenic-like |
| chr9_-_15424639 | 0.26 |
ENSDART00000124346
|
fn1a
|
fibronectin 1a |
| chr3_-_43646733 | 0.23 |
ENSDART00000180959
|
axin1
|
axin 1 |
| chr7_-_38570878 | 0.22 |
ENSDART00000139187
ENSDART00000134570 ENSDART00000041055 |
celf1
|
cugbp, Elav-like family member 1 |
| chr9_+_33334501 | 0.21 |
ENSDART00000006867
|
ddx3a
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3a |
| chr7_-_38570299 | 0.21 |
ENSDART00000143815
|
celf1
|
cugbp, Elav-like family member 1 |
| chr25_-_32363341 | 0.21 |
ENSDART00000153892
ENSDART00000114385 |
cep152
|
centrosomal protein 152 |
| chr21_+_21279159 | 0.18 |
ENSDART00000148346
|
itpkca
|
inositol-trisphosphate 3-kinase Ca |
| chr2_-_21847935 | 0.18 |
ENSDART00000003940
|
rab2a
|
RAB2A, member RAS oncogene family |
| chr14_+_31509922 | 0.17 |
ENSDART00000124499
|
hprt1
|
hypoxanthine phosphoribosyltransferase 1 |
| chr21_+_21205667 | 0.16 |
ENSDART00000058311
|
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr10_+_6383270 | 0.16 |
ENSDART00000170548
|
zgc:114200
|
zgc:114200 |
| chr16_+_54209504 | 0.15 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr5_-_7513082 | 0.15 |
ENSDART00000158913
|
bmpr1ba
|
bone morphogenetic protein receptor, type IBa |
| chr20_-_54014373 | 0.15 |
ENSDART00000152934
|
si:dkey-241l7.6
|
si:dkey-241l7.6 |
| chr16_-_21620947 | 0.15 |
ENSDART00000115011
ENSDART00000183125 ENSDART00000188856 ENSDART00000189460 |
ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr25_+_22017182 | 0.14 |
ENSDART00000156517
|
si:dkey-217l24.1
|
si:dkey-217l24.1 |
| chr1_+_58990121 | 0.13 |
ENSDART00000171654
|
CABZ01083166.1
|
|
| chr21_-_34926619 | 0.13 |
ENSDART00000065337
ENSDART00000192740 |
kif20a
|
kinesin family member 20A |
| chr13_+_16521898 | 0.12 |
ENSDART00000122557
|
kcnma1a
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1a |
| chr7_+_19482877 | 0.12 |
ENSDART00000077868
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr7_+_1442059 | 0.11 |
ENSDART00000173391
|
si:cabz01090193.1
|
si:cabz01090193.1 |
| chr4_-_73548389 | 0.10 |
ENSDART00000174327
ENSDART00000150753 ENSDART00000170775 |
si:ch73-266f23.1
|
si:ch73-266f23.1 |
| chr21_+_26748141 | 0.10 |
ENSDART00000169025
|
pcxa
|
pyruvate carboxylase a |
| chr13_+_36585399 | 0.09 |
ENSDART00000030211
|
gmfb
|
glia maturation factor, beta |
| chr11_-_24526503 | 0.09 |
ENSDART00000181252
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr12_+_5708400 | 0.07 |
ENSDART00000017191
|
dlx3b
|
distal-less homeobox 3b |
| chr18_+_18104235 | 0.07 |
ENSDART00000145342
|
cbln1
|
cerebellin 1 precursor |
| chr7_+_1521834 | 0.07 |
ENSDART00000174007
|
si:cabz01102082.1
|
si:cabz01102082.1 |
| chr12_-_8958353 | 0.07 |
ENSDART00000041728
|
cyp26a1
|
cytochrome P450, family 26, subfamily A, polypeptide 1 |
| chr23_-_1017428 | 0.06 |
ENSDART00000110588
ENSDART00000183158 |
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr10_-_43964028 | 0.06 |
ENSDART00000009134
ENSDART00000133450 |
sept5b
|
septin 5b |
| chr6_+_7322587 | 0.06 |
ENSDART00000065500
|
abcc4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr8_+_24281512 | 0.06 |
ENSDART00000062845
|
mmp9
|
matrix metallopeptidase 9 |
| chr10_-_5135788 | 0.06 |
ENSDART00000108587
ENSDART00000138537 |
sec31a
|
SEC31 homolog A, COPII coat complex component |
| chr22_-_9677264 | 0.06 |
ENSDART00000181737
|
si:dkey-286j17.4
|
si:dkey-286j17.4 |
| chr25_+_6471401 | 0.05 |
ENSDART00000132927
|
snx33
|
sorting nexin 33 |
| chr8_+_26329482 | 0.05 |
ENSDART00000048676
|
prkar2aa
|
protein kinase, cAMP-dependent, regulatory, type II, alpha A |
| chr25_-_20870900 | 0.04 |
ENSDART00000171288
|
magi2b
|
membrane associated guanylate kinase, WW and PDZ domain containing 2b |
| chr9_+_44721808 | 0.03 |
ENSDART00000190578
|
nckap1
|
NCK-associated protein 1 |
| chr9_+_22764235 | 0.03 |
ENSDART00000090875
|
tnfaip6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr23_-_27701361 | 0.03 |
ENSDART00000186688
ENSDART00000183985 |
dnajc22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr23_+_39461989 | 0.03 |
ENSDART00000184761
|
mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr11_-_35975026 | 0.03 |
ENSDART00000186219
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr23_-_1017605 | 0.03 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr15_-_18213515 | 0.02 |
ENSDART00000101635
|
btr21
|
bloodthirsty-related gene family, member 21 |
| chr7_+_19482084 | 0.02 |
ENSDART00000173873
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr17_-_26868169 | 0.02 |
ENSDART00000157204
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr6_-_19035749 | 0.02 |
ENSDART00000187714
|
sept9b
|
septin 9b |
| chr6_+_102506 | 0.02 |
ENSDART00000172678
|
ldlrb
|
low density lipoprotein receptor b |
| chr2_-_30324297 | 0.02 |
ENSDART00000099078
|
jph1b
|
junctophilin 1b |
| chr1_-_43987873 | 0.02 |
ENSDART00000108821
|
CR385050.1
|
|
| chr14_+_10450216 | 0.02 |
ENSDART00000134814
|
cysltr1
|
cysteinyl leukotriene receptor 1 |
| chr25_+_16945348 | 0.02 |
ENSDART00000016591
|
fgf6a
|
fibroblast growth factor 6a |
| chr4_+_54439143 | 0.02 |
ENSDART00000170168
|
si:ch211-133h13.1
|
si:ch211-133h13.1 |
| chr3_+_16455749 | 0.01 |
ENSDART00000090767
|
kcnj12b
|
potassium inwardly-rectifying channel, subfamily J, member 12b |
| chr4_-_20115570 | 0.01 |
ENSDART00000134528
|
si:dkey-159a18.1
|
si:dkey-159a18.1 |
| chr17_-_27972307 | 0.01 |
ENSDART00000062130
|
ptafr
|
platelet-activating factor receptor |
| chr22_-_17953529 | 0.01 |
ENSDART00000135716
|
ncanb
|
neurocan b |
| chr12_+_30360579 | 0.01 |
ENSDART00000152900
|
si:ch211-225b10.3
|
si:ch211-225b10.3 |
| chr12_+_30360184 | 0.01 |
ENSDART00000190718
|
si:ch211-225b10.3
|
si:ch211-225b10.3 |
| chr2_-_31800521 | 0.01 |
ENSDART00000112763
|
retreg1
|
reticulophagy regulator 1 |
| chr18_+_33212589 | 0.01 |
ENSDART00000028134
|
olfcd1
|
olfactory receptor C family, d1 |
| chr6_-_2171818 | 0.01 |
ENSDART00000181082
ENSDART00000187556 |
PXMP4
|
wu:fe02h09 |
| chr4_+_58667348 | 0.01 |
ENSDART00000186596
ENSDART00000180673 |
CR388148.2
|
|
| chr6_+_38381957 | 0.01 |
ENSDART00000087300
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr7_-_61842593 | 0.01 |
ENSDART00000023770
|
HTRA3
|
HtrA serine peptidase 3 |
| chr4_-_72513569 | 0.01 |
ENSDART00000174130
|
CR788316.4
|
|
| chr18_+_24919614 | 0.01 |
ENSDART00000008638
|
rgma
|
repulsive guidance molecule family member a |
| chr23_-_9965148 | 0.01 |
ENSDART00000137308
|
plxnb1a
|
plexin b1a |
| chr25_+_21180695 | 0.00 |
ENSDART00000012923
|
erc1a
|
ELKS/RAB6-interacting/CAST family member 1a |
| chr5_-_15948833 | 0.00 |
ENSDART00000051649
ENSDART00000124467 |
xbp1
|
X-box binding protein 1 |
| chr3_-_33030727 | 0.00 |
ENSDART00000157591
ENSDART00000193200 |
CT583642.1
|
|
| chr12_-_21834611 | 0.00 |
ENSDART00000179553
|
thrab
|
thyroid hormone receptor alpha b |
| chr9_-_48864942 | 0.00 |
ENSDART00000113855
ENSDART00000189064 |
nostrin
|
nitric oxide synthase trafficking |
Network of associatons between targets according to the STRING database.
First level regulatory network of tfap2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0061113 | endodermal digestive tract morphogenesis(GO:0061031) pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.3 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.2 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 | 0.2 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.1 | 0.2 | GO:0032263 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.0 | 0.3 | GO:0003190 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.3 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) acute-phase response(GO:0006953) establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:0030910 | olfactory placode formation(GO:0030910) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0002631 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.0 | 0.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 0.0 | GO:0019417 | sulfur oxidation(GO:0019417) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.7 | GO:0045495 | pole plasm(GO:0045495) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.1 | 0.2 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.1 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.3 | GO:0001948 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.0 | 0.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.0 | GO:0070699 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.2 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |