Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
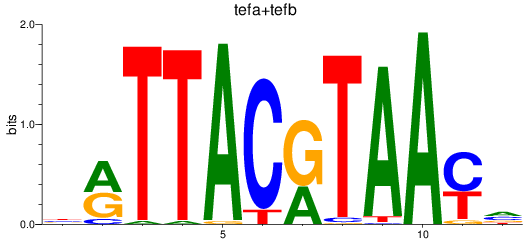
Results for tefa+tefb
Z-value: 0.93
Transcription factors associated with tefa+tefb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tefa
|
ENSDARG00000039117 | TEF transcription factor, PAR bZIP family member a |
|
tefb
|
ENSDARG00000098103 | TEF transcription factor, PAR bZIP family member b |
|
tefa
|
ENSDARG00000109532 | TEF transcription factor, PAR bZIP family member a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tefb | dr11_v1_chr3_-_5067585_5067585 | 0.99 | 3.2e-14 | Click! |
| tefa | dr11_v1_chr12_-_19153177_19153177 | 0.64 | 4.1e-03 | Click! |
Activity profile of tefa+tefb motif
Sorted Z-values of tefa+tefb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_22974019 | 4.74 |
ENSDART00000147157
ENSDART00000020434 |
brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr3_+_7763114 | 1.73 |
ENSDART00000057434
|
hook2
|
hook microtubule-tethering protein 2 |
| chr5_-_69212184 | 1.67 |
ENSDART00000053963
|
mat2ab
|
methionine adenosyltransferase II, alpha b |
| chr14_+_21699129 | 1.61 |
ENSDART00000073707
|
stx3a
|
syntaxin 3A |
| chr7_+_38380135 | 1.48 |
ENSDART00000174005
|
rhpn2
|
rhophilin, Rho GTPase binding protein 2 |
| chr18_-_39288894 | 1.44 |
ENSDART00000186216
|
mapk6
|
mitogen-activated protein kinase 6 |
| chr22_+_10606863 | 1.38 |
ENSDART00000147975
|
rad54l2
|
RAD54 like 2 |
| chr3_-_26184018 | 1.35 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr5_-_69716501 | 1.33 |
ENSDART00000158956
|
mob1a
|
MOB kinase activator 1A |
| chr15_+_24737599 | 1.31 |
ENSDART00000078024
|
crk
|
v-crk avian sarcoma virus CT10 oncogene homolog |
| chr1_+_16600690 | 1.28 |
ENSDART00000162164
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr14_+_21699414 | 1.25 |
ENSDART00000169942
|
stx3a
|
syntaxin 3A |
| chr8_-_11324143 | 1.24 |
ENSDART00000008215
|
pip5k1bb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta b |
| chr7_-_40959667 | 1.22 |
ENSDART00000084070
|
rbm33a
|
RNA binding motif protein 33a |
| chr20_+_38201644 | 1.15 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr1_+_19764995 | 1.15 |
ENSDART00000138276
|
si:ch211-42i9.8
|
si:ch211-42i9.8 |
| chr8_+_21225064 | 1.13 |
ENSDART00000129210
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr19_+_7549854 | 1.11 |
ENSDART00000138866
ENSDART00000151758 |
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr21_+_43178831 | 1.10 |
ENSDART00000151512
|
aff4
|
AF4/FMR2 family, member 4 |
| chr23_+_33963619 | 1.06 |
ENSDART00000140666
ENSDART00000084792 |
plpbp
|
pyridoxal phosphate binding protein |
| chr5_-_65153731 | 1.01 |
ENSDART00000171024
|
sh2d3cb
|
SH2 domain containing 3Cb |
| chr4_+_5341592 | 1.00 |
ENSDART00000123375
ENSDART00000067371 |
zgc:113263
|
zgc:113263 |
| chr3_+_43373867 | 0.99 |
ENSDART00000159455
ENSDART00000172425 |
zfand2a
|
zinc finger, AN1-type domain 2A |
| chr16_-_35427060 | 0.97 |
ENSDART00000172294
|
ctps1b
|
CTP synthase 1b |
| chr3_-_26183699 | 0.97 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr23_+_9522781 | 0.96 |
ENSDART00000136486
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr14_+_3507326 | 0.92 |
ENSDART00000159326
|
gstp1
|
glutathione S-transferase pi 1 |
| chr16_-_6849754 | 0.88 |
ENSDART00000149206
ENSDART00000149778 |
mbpb
|
myelin basic protein b |
| chr20_-_36617313 | 0.87 |
ENSDART00000172395
ENSDART00000152856 |
enah
|
enabled homolog (Drosophila) |
| chr1_-_354115 | 0.87 |
ENSDART00000141590
ENSDART00000098627 |
pros1
|
protein S |
| chr13_+_30035253 | 0.86 |
ENSDART00000181303
ENSDART00000057525 ENSDART00000136622 |
dnajb12a
|
DnaJ (Hsp40) homolog, subfamily B, member 12a |
| chr18_-_14879135 | 0.85 |
ENSDART00000099701
|
selenoo1
|
selenoprotein O1 |
| chr21_-_32467799 | 0.85 |
ENSDART00000007675
ENSDART00000133099 |
zgc:123105
|
zgc:123105 |
| chr16_+_39146696 | 0.85 |
ENSDART00000121756
ENSDART00000084381 |
sybu
|
syntabulin (syntaxin-interacting) |
| chr23_-_21763598 | 0.84 |
ENSDART00000145408
|
vps13d
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr17_-_23709347 | 0.84 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr21_-_32467099 | 0.79 |
ENSDART00000186354
|
zgc:123105
|
zgc:123105 |
| chr10_+_18911152 | 0.78 |
ENSDART00000030205
|
bnip3lb
|
BCL2 interacting protein 3 like b |
| chr12_-_26383242 | 0.70 |
ENSDART00000152941
|
usp54b
|
ubiquitin specific peptidase 54b |
| chr16_-_19568388 | 0.68 |
ENSDART00000141616
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr14_-_30905288 | 0.67 |
ENSDART00000173449
ENSDART00000173451 |
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr12_-_23365737 | 0.67 |
ENSDART00000170376
|
mpp7a
|
membrane protein, palmitoylated 7a (MAGUK p55 subfamily member 7) |
| chr16_+_40954481 | 0.64 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr7_+_38278860 | 0.62 |
ENSDART00000016265
|
lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr11_-_40147032 | 0.61 |
ENSDART00000052918
|
si:dkey-264d12.4
|
si:dkey-264d12.4 |
| chr9_-_9225980 | 0.57 |
ENSDART00000180301
|
cbsb
|
cystathionine-beta-synthase b |
| chr14_+_1170968 | 0.56 |
ENSDART00000125203
ENSDART00000193575 |
hopx
|
HOP homeobox |
| chr14_+_23717165 | 0.56 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr18_+_25546227 | 0.56 |
ENSDART00000085824
|
pex11a
|
peroxisomal biogenesis factor 11 alpha |
| chr19_-_45650994 | 0.55 |
ENSDART00000163504
|
trps1
|
trichorhinophalangeal syndrome I |
| chr22_+_10606573 | 0.53 |
ENSDART00000192638
|
rad54l2
|
RAD54 like 2 |
| chr9_-_13871935 | 0.53 |
ENSDART00000146597
|
raph1a
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1a |
| chr23_-_18981595 | 0.52 |
ENSDART00000147617
|
bcl2l1
|
bcl2-like 1 |
| chr24_-_23784701 | 0.51 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr25_-_12803723 | 0.51 |
ENSDART00000158787
|
ca5a
|
carbonic anhydrase Va |
| chr9_-_12888082 | 0.51 |
ENSDART00000133135
ENSDART00000134415 |
si:ch211-167j6.3
|
si:ch211-167j6.3 |
| chr20_+_23083800 | 0.48 |
ENSDART00000132093
|
usp46
|
ubiquitin specific peptidase 46 |
| chr6_+_16468776 | 0.45 |
ENSDART00000109151
ENSDART00000114667 |
zgc:161969
|
zgc:161969 |
| chr20_-_44576949 | 0.39 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr5_-_54395488 | 0.39 |
ENSDART00000160781
|
zmynd19
|
zinc finger, MYND-type containing 19 |
| chr23_+_19590006 | 0.39 |
ENSDART00000021231
|
slmapb
|
sarcolemma associated protein b |
| chr10_-_25591194 | 0.34 |
ENSDART00000131640
|
tiam1a
|
T cell lymphoma invasion and metastasis 1a |
| chr17_-_8268406 | 0.28 |
ENSDART00000149873
ENSDART00000064668 ENSDART00000148403 |
ahi1
|
Abelson helper integration site 1 |
| chr22_+_15973122 | 0.27 |
ENSDART00000144545
|
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr18_-_2549198 | 0.25 |
ENSDART00000186516
|
CABZ01070631.1
|
|
| chr14_+_34971554 | 0.25 |
ENSDART00000184271
|
rnf145a
|
ring finger protein 145a |
| chr4_+_21866851 | 0.25 |
ENSDART00000177640
|
acss3
|
acyl-CoA synthetase short chain family member 3 |
| chr15_+_21882419 | 0.24 |
ENSDART00000157216
|
si:dkey-103g5.4
|
si:dkey-103g5.4 |
| chr5_+_69808763 | 0.24 |
ENSDART00000143482
|
fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr17_+_49281597 | 0.23 |
ENSDART00000155599
|
zgc:113176
|
zgc:113176 |
| chr25_-_7887274 | 0.23 |
ENSDART00000157276
|
si:ch73-151m17.5
|
si:ch73-151m17.5 |
| chr20_+_1121458 | 0.19 |
ENSDART00000064472
|
pnrc1
|
proline-rich nuclear receptor coactivator 1 |
| chr9_+_46644633 | 0.19 |
ENSDART00000160285
|
slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr24_-_39772045 | 0.19 |
ENSDART00000087441
|
GFOD1
|
si:ch211-276f18.2 |
| chr3_-_21348478 | 0.17 |
ENSDART00000114906
|
fam171a2a
|
family with sequence similarity 171, member A2a |
| chr15_-_31419805 | 0.16 |
ENSDART00000060111
|
or111-11
|
odorant receptor, family D, subfamily 111, member 11 |
| chr10_+_22771176 | 0.16 |
ENSDART00000192046
|
tmem88a
|
transmembrane protein 88 a |
| chr25_+_34014523 | 0.15 |
ENSDART00000182856
|
anxa2a
|
annexin A2a |
| chr5_+_26121393 | 0.10 |
ENSDART00000002221
|
bco2l
|
beta-carotene 15, 15-dioxygenase 2, like |
| chr21_+_21374277 | 0.09 |
ENSDART00000079431
|
rtn2b
|
reticulon 2b |
| chr22_-_13165186 | 0.08 |
ENSDART00000105762
|
ahr2
|
aryl hydrocarbon receptor 2 |
| chr4_-_12914163 | 0.06 |
ENSDART00000140002
ENSDART00000145917 ENSDART00000141355 ENSDART00000067135 |
msrb3
|
methionine sulfoxide reductase B3 |
| chr21_-_39566854 | 0.05 |
ENSDART00000020174
|
dynll2b
|
dynein, light chain, LC8-type 2b |
| chr3_+_24094581 | 0.04 |
ENSDART00000138270
ENSDART00000131509 |
copz2
|
coatomer protein complex, subunit zeta 2 |
| chr24_+_18948665 | 0.03 |
ENSDART00000106186
|
prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr24_+_28561038 | 0.03 |
ENSDART00000147063
|
abca4a
|
ATP-binding cassette, sub-family A (ABC1), member 4a |
| chr6_-_48347724 | 0.02 |
ENSDART00000046973
|
capza1a
|
capping protein (actin filament) muscle Z-line, alpha 1a |
| chr4_+_21867522 | 0.02 |
ENSDART00000140400
|
acss3
|
acyl-CoA synthetase short chain family member 3 |
| chr9_-_44953664 | 0.02 |
ENSDART00000188558
ENSDART00000185210 |
vil1
|
villin 1 |
| chr9_-_44953349 | 0.01 |
ENSDART00000135156
|
vil1
|
villin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tefa+tefb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.9 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.3 | 0.9 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.3 | 0.9 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.3 | 0.8 | GO:0050427 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.2 | 1.0 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.2 | 1.7 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 0.7 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.2 | 0.8 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.2 | 0.6 | GO:0050666 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.2 | 1.1 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 0.8 | GO:0045822 | negative regulation of heart contraction(GO:0045822) negative regulation of blood circulation(GO:1903523) |
| 0.1 | 0.8 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.6 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 1.0 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.1 | 1.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.5 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.1 | 0.2 | GO:1901295 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.0 | 1.0 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 4.7 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.9 | GO:0051788 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.5 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.1 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.0 | 1.7 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 1.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.9 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.6 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 2.3 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 0.2 | GO:0086001 | cardiac muscle cell action potential(GO:0086001) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 1.0 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.1 | 1.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 2.9 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 1.0 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 1.1 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.3 | 0.8 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.2 | 1.0 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.6 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.1 | 0.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.9 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.9 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.9 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 0.3 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.0 | 2.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.5 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 1.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 1.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 1.2 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 1.0 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 1.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.1 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.3 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 0.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 0.5 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 1.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 0.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |